
bacterium 0.1xD8-71
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
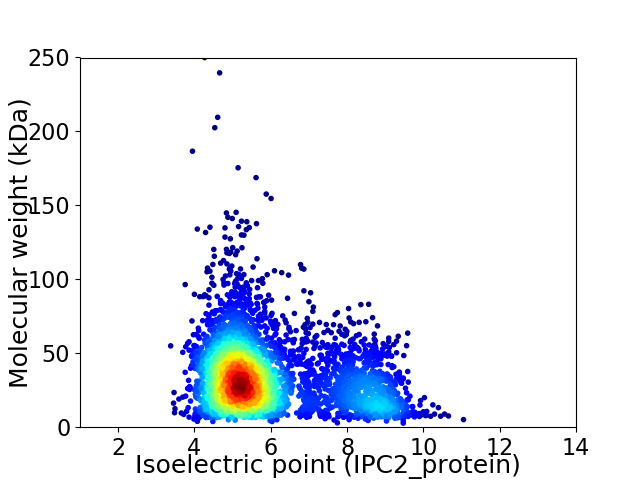
Virtual 2D-PAGE plot for 3809 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3A8ZI62|A0A3A8ZI62_9BACT Sugar ABC transporter permease OS=bacterium 0.1xD8-71 OX=2320099 GN=D7V83_16380 PE=3 SV=1
MM1 pKa = 7.54KK2 pKa = 10.17LKK4 pKa = 10.51KK5 pKa = 10.46VLALTLAAAMTFSLAACGGSDD26 pKa = 3.57EE27 pKa = 4.87GAAEE31 pKa = 4.32APATTQEE38 pKa = 4.22TGGSEE43 pKa = 3.82EE44 pKa = 4.09AQAPEE49 pKa = 4.09AEE51 pKa = 4.37EE52 pKa = 4.48AEE54 pKa = 4.51PEE56 pKa = 4.14AEE58 pKa = 4.18APAAAGVTYY67 pKa = 10.95ASINLGEE74 pKa = 4.71DD75 pKa = 3.56YY76 pKa = 11.16KK77 pKa = 11.65DD78 pKa = 3.08LTTTIKK84 pKa = 10.53FIHH87 pKa = 6.65HH88 pKa = 5.44KK89 pKa = 8.52TDD91 pKa = 3.41RR92 pKa = 11.84EE93 pKa = 4.03EE94 pKa = 4.86DD95 pKa = 3.25GTMASLVSKK104 pKa = 10.61FNEE107 pKa = 4.18VYY109 pKa = 10.73PNITVEE115 pKa = 4.28TEE117 pKa = 4.01GITDD121 pKa = 3.69YY122 pKa = 11.64QEE124 pKa = 5.59DD125 pKa = 3.9SLLRR129 pKa = 11.84LSTGDD134 pKa = 2.78WGDD137 pKa = 2.91IMFIPAIDD145 pKa = 3.67KK146 pKa = 11.33AEE148 pKa = 4.07LSTYY152 pKa = 8.78FIPYY156 pKa = 7.88GTVSEE161 pKa = 4.25MSGLVNFADD170 pKa = 3.07EE171 pKa = 3.95WMYY174 pKa = 11.94DD175 pKa = 3.55DD176 pKa = 3.42MCYY179 pKa = 10.64GVGYY183 pKa = 9.22MGNAQGLLYY192 pKa = 10.07NKK194 pKa = 10.03KK195 pKa = 8.82VFADD199 pKa = 3.44AGVTQLPKK207 pKa = 10.67TPDD210 pKa = 3.06EE211 pKa = 5.17FIAALQAIKK220 pKa = 10.77DD221 pKa = 3.84NTDD224 pKa = 4.57AIPLYY229 pKa = 10.26TNYY232 pKa = 10.71AAGWTMGGQWDD243 pKa = 4.2AFLGGITTGDD253 pKa = 3.91EE254 pKa = 3.36TWMNQKK260 pKa = 9.42FVHH263 pKa = 5.33TAEE266 pKa = 4.27PFKK269 pKa = 11.42DD270 pKa = 3.49NGDD273 pKa = 3.53GTGAYY278 pKa = 9.94ALYY281 pKa = 10.2KK282 pKa = 10.0ILYY285 pKa = 9.01DD286 pKa = 3.65AVEE289 pKa = 3.94QGLIEE294 pKa = 5.49DD295 pKa = 5.46DD296 pKa = 3.88YY297 pKa = 10.81TTTDD301 pKa = 3.01WEE303 pKa = 4.91GCKK306 pKa = 10.32PMINNGEE313 pKa = 4.19IGCMVLGSWAIAQMKK328 pKa = 9.56AAGDD332 pKa = 3.73NPDD335 pKa = 5.11DD336 pKa = 3.69IGYY339 pKa = 8.2MPFPITVGGKK349 pKa = 9.24QYY351 pKa = 10.3ATAGPDD357 pKa = 3.18YY358 pKa = 11.23SFGINANASEE368 pKa = 4.59DD369 pKa = 3.73NQAAAMVFVKK379 pKa = 10.09WLVEE383 pKa = 3.77EE384 pKa = 5.1SGWCDD389 pKa = 3.52FEE391 pKa = 5.99GGYY394 pKa = 9.61PISKK398 pKa = 8.26TAPTSFVFDD407 pKa = 3.29GVEE410 pKa = 3.71IVRR413 pKa = 11.84NIPALEE419 pKa = 4.19GEE421 pKa = 4.06EE422 pKa = 5.43DD423 pKa = 4.0YY424 pKa = 11.54LNEE427 pKa = 3.92MNAEE431 pKa = 4.18SEE433 pKa = 4.36LSFNAGGDD441 pKa = 3.38AKK443 pKa = 10.31VQRR446 pKa = 11.84IVEE449 pKa = 4.09AAATGAEE456 pKa = 4.24TYY458 pKa = 11.1DD459 pKa = 5.58DD460 pKa = 5.71IMADD464 pKa = 3.09WTAAWNAAQEE474 pKa = 4.31EE475 pKa = 4.97CGVEE479 pKa = 3.91VLYY482 pKa = 11.07
MM1 pKa = 7.54KK2 pKa = 10.17LKK4 pKa = 10.51KK5 pKa = 10.46VLALTLAAAMTFSLAACGGSDD26 pKa = 3.57EE27 pKa = 4.87GAAEE31 pKa = 4.32APATTQEE38 pKa = 4.22TGGSEE43 pKa = 3.82EE44 pKa = 4.09AQAPEE49 pKa = 4.09AEE51 pKa = 4.37EE52 pKa = 4.48AEE54 pKa = 4.51PEE56 pKa = 4.14AEE58 pKa = 4.18APAAAGVTYY67 pKa = 10.95ASINLGEE74 pKa = 4.71DD75 pKa = 3.56YY76 pKa = 11.16KK77 pKa = 11.65DD78 pKa = 3.08LTTTIKK84 pKa = 10.53FIHH87 pKa = 6.65HH88 pKa = 5.44KK89 pKa = 8.52TDD91 pKa = 3.41RR92 pKa = 11.84EE93 pKa = 4.03EE94 pKa = 4.86DD95 pKa = 3.25GTMASLVSKK104 pKa = 10.61FNEE107 pKa = 4.18VYY109 pKa = 10.73PNITVEE115 pKa = 4.28TEE117 pKa = 4.01GITDD121 pKa = 3.69YY122 pKa = 11.64QEE124 pKa = 5.59DD125 pKa = 3.9SLLRR129 pKa = 11.84LSTGDD134 pKa = 2.78WGDD137 pKa = 2.91IMFIPAIDD145 pKa = 3.67KK146 pKa = 11.33AEE148 pKa = 4.07LSTYY152 pKa = 8.78FIPYY156 pKa = 7.88GTVSEE161 pKa = 4.25MSGLVNFADD170 pKa = 3.07EE171 pKa = 3.95WMYY174 pKa = 11.94DD175 pKa = 3.55DD176 pKa = 3.42MCYY179 pKa = 10.64GVGYY183 pKa = 9.22MGNAQGLLYY192 pKa = 10.07NKK194 pKa = 10.03KK195 pKa = 8.82VFADD199 pKa = 3.44AGVTQLPKK207 pKa = 10.67TPDD210 pKa = 3.06EE211 pKa = 5.17FIAALQAIKK220 pKa = 10.77DD221 pKa = 3.84NTDD224 pKa = 4.57AIPLYY229 pKa = 10.26TNYY232 pKa = 10.71AAGWTMGGQWDD243 pKa = 4.2AFLGGITTGDD253 pKa = 3.91EE254 pKa = 3.36TWMNQKK260 pKa = 9.42FVHH263 pKa = 5.33TAEE266 pKa = 4.27PFKK269 pKa = 11.42DD270 pKa = 3.49NGDD273 pKa = 3.53GTGAYY278 pKa = 9.94ALYY281 pKa = 10.2KK282 pKa = 10.0ILYY285 pKa = 9.01DD286 pKa = 3.65AVEE289 pKa = 3.94QGLIEE294 pKa = 5.49DD295 pKa = 5.46DD296 pKa = 3.88YY297 pKa = 10.81TTTDD301 pKa = 3.01WEE303 pKa = 4.91GCKK306 pKa = 10.32PMINNGEE313 pKa = 4.19IGCMVLGSWAIAQMKK328 pKa = 9.56AAGDD332 pKa = 3.73NPDD335 pKa = 5.11DD336 pKa = 3.69IGYY339 pKa = 8.2MPFPITVGGKK349 pKa = 9.24QYY351 pKa = 10.3ATAGPDD357 pKa = 3.18YY358 pKa = 11.23SFGINANASEE368 pKa = 4.59DD369 pKa = 3.73NQAAAMVFVKK379 pKa = 10.09WLVEE383 pKa = 3.77EE384 pKa = 5.1SGWCDD389 pKa = 3.52FEE391 pKa = 5.99GGYY394 pKa = 9.61PISKK398 pKa = 8.26TAPTSFVFDD407 pKa = 3.29GVEE410 pKa = 3.71IVRR413 pKa = 11.84NIPALEE419 pKa = 4.19GEE421 pKa = 4.06EE422 pKa = 5.43DD423 pKa = 4.0YY424 pKa = 11.54LNEE427 pKa = 3.92MNAEE431 pKa = 4.18SEE433 pKa = 4.36LSFNAGGDD441 pKa = 3.38AKK443 pKa = 10.31VQRR446 pKa = 11.84IVEE449 pKa = 4.09AAATGAEE456 pKa = 4.24TYY458 pKa = 11.1DD459 pKa = 5.58DD460 pKa = 5.71IMADD464 pKa = 3.09WTAAWNAAQEE474 pKa = 4.31EE475 pKa = 4.97CGVEE479 pKa = 3.91VLYY482 pKa = 11.07
Molecular weight: 52.06 kDa
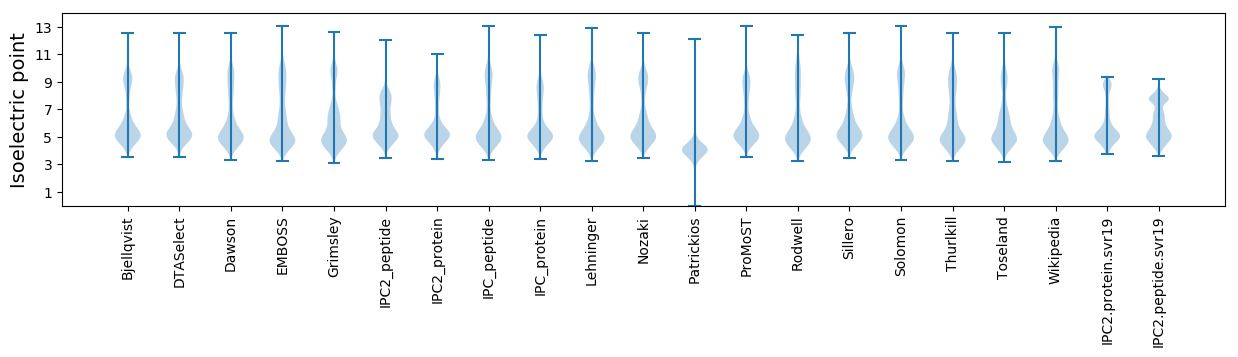
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3A9A4F8|A0A3A9A4F8_9BACT Thioredoxin OS=bacterium 0.1xD8-71 OX=2320099 GN=D7V83_10870 PE=4 SV=1
MM1 pKa = 7.6KK2 pKa = 8.56MTYY5 pKa = 8.56QPKK8 pKa = 9.32KK9 pKa = 7.54RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.15GGRR28 pKa = 11.84KK29 pKa = 8.78VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.6KK2 pKa = 8.56MTYY5 pKa = 8.56QPKK8 pKa = 9.32KK9 pKa = 7.54RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.15GGRR28 pKa = 11.84KK29 pKa = 8.78VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1197674 |
25 |
2354 |
314.4 |
35.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.455 ± 0.045 | 1.55 ± 0.013 |
5.714 ± 0.031 | 7.87 ± 0.039 |
4.025 ± 0.029 | 7.08 ± 0.036 |
1.76 ± 0.015 | 7.195 ± 0.037 |
6.205 ± 0.037 | 8.982 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.274 ± 0.018 | 4.147 ± 0.024 |
3.146 ± 0.02 | 3.551 ± 0.026 |
4.861 ± 0.03 | 5.53 ± 0.032 |
5.17 ± 0.029 | 6.982 ± 0.033 |
1.008 ± 0.014 | 4.493 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
