
Flavobacteriaceae bacterium R38
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; unclassified Flavobacteriaceae
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
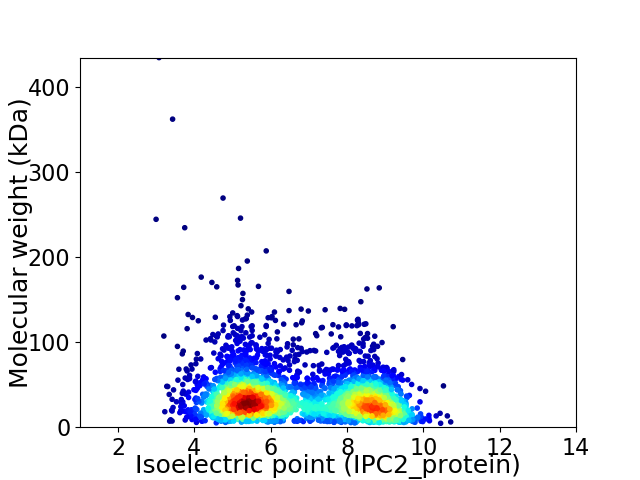
Virtual 2D-PAGE plot for 3541 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
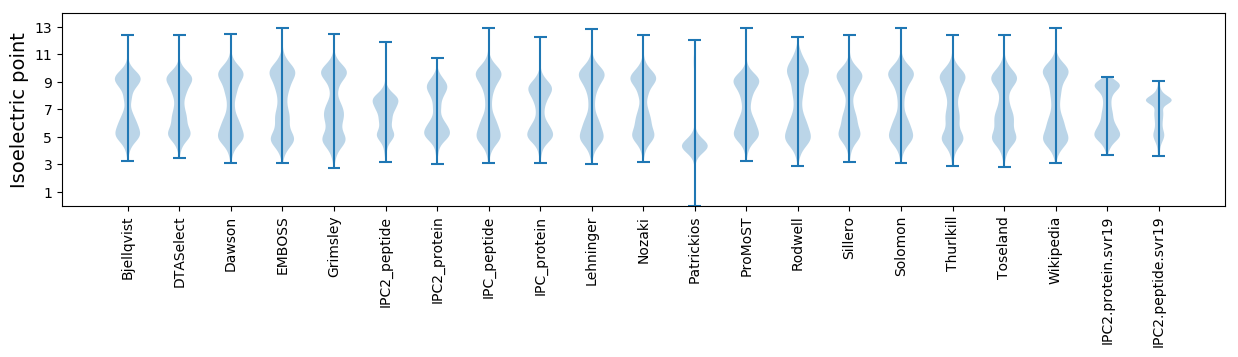
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6L9EVR1|A0A6L9EVR1_9FLAO DUF2911 domain-containing protein OS=Flavobacteriaceae bacterium R38 OX=2697054 GN=GTQ40_01500 PE=4 SV=1
MM1 pKa = 7.68RR2 pKa = 11.84FISVFIFLIFFCFSNKK18 pKa = 7.88TFAQNIPPTILVEE31 pKa = 4.21NEE33 pKa = 3.72PFYY36 pKa = 11.47CPLSRR41 pKa = 11.84QNIVTAFDD49 pKa = 4.66IIDD52 pKa = 4.18PDD54 pKa = 3.79DD55 pKa = 4.12TEE57 pKa = 4.47VEE59 pKa = 4.03AFFIQISVGYY69 pKa = 10.41DD70 pKa = 2.9EE71 pKa = 5.98GQDD74 pKa = 3.72FLEE77 pKa = 4.43LTNSDD82 pKa = 3.88FHH84 pKa = 8.2PEE86 pKa = 4.01IVAIWNSEE94 pKa = 3.9EE95 pKa = 4.13GRR97 pKa = 11.84LTLEE101 pKa = 4.11GVGGIMIDD109 pKa = 3.45YY110 pKa = 9.78DD111 pKa = 4.39AIIAAVNDD119 pKa = 3.21IVFFNTDD126 pKa = 3.67PDD128 pKa = 3.75NTTSKK133 pKa = 10.56SFSYY137 pKa = 10.53TIGDD141 pKa = 3.93ANFLPSTGHH150 pKa = 6.87FYY152 pKa = 11.09QFIPDD157 pKa = 4.34IGISWIAAEE166 pKa = 4.08QAASNLTFFGLQGYY180 pKa = 8.27LATLTSAEE188 pKa = 4.13EE189 pKa = 3.79AQFAGNQITGNGWIGGSDD207 pKa = 3.25AAEE210 pKa = 3.89EE211 pKa = 4.69GVWRR215 pKa = 11.84WVTGPEE221 pKa = 3.55NGMVFWNGGVNGSTPNFANWNTGEE245 pKa = 4.01PNQLGNEE252 pKa = 4.3DD253 pKa = 3.98YY254 pKa = 11.79AHH256 pKa = 6.26ITSPGIGIDD265 pKa = 4.03GSWNDD270 pKa = 4.2LRR272 pKa = 11.84NEE274 pKa = 4.39GDD276 pKa = 3.18PSGPFQSMGYY286 pKa = 7.97VVEE289 pKa = 4.18YY290 pKa = 10.66GGLPGDD296 pKa = 3.55PVINISASTTFMIPTILSVSNDD318 pKa = 3.04INSCGPVEE326 pKa = 4.39VDD328 pKa = 3.97LTATINGEE336 pKa = 4.13RR337 pKa = 11.84IFWFASEE344 pKa = 4.1TSDD347 pKa = 4.59VILHH351 pKa = 5.97TGIPFTVNVEE361 pKa = 3.91NTTTFWVLASVDD373 pKa = 3.46GCLTGVRR380 pKa = 11.84TPVTVMIDD388 pKa = 3.53GLPSINNASLTQCDD402 pKa = 3.28VDD404 pKa = 4.24GVSDD408 pKa = 4.3GSTVFNLNEE417 pKa = 4.04ARR419 pKa = 11.84TALIGDD425 pKa = 4.0ITNVSIQFFTSLIDD439 pKa = 3.62AQNATNAVNSNVFRR453 pKa = 11.84NTVNPQTLFVRR464 pKa = 11.84ANDD467 pKa = 3.53NVSACFSIAEE477 pKa = 4.24LVLDD481 pKa = 4.42VSTTQVNNATLEE493 pKa = 4.19VCDD496 pKa = 3.94TDD498 pKa = 4.21GNEE501 pKa = 4.68DD502 pKa = 3.74GLHH505 pKa = 6.65LFTLSDD511 pKa = 4.06ADD513 pKa = 4.92DD514 pKa = 5.01EE515 pKa = 4.53ILNGLTNNGDD525 pKa = 3.48FDD527 pKa = 3.76IVYY530 pKa = 10.18YY531 pKa = 11.03ASTEE535 pKa = 4.1DD536 pKa = 5.22ALLEE540 pKa = 4.08QNPLNNDD547 pKa = 3.61YY548 pKa = 11.9ANVTPGNQIIYY559 pKa = 10.87ARR561 pKa = 11.84IEE563 pKa = 4.2DD564 pKa = 3.85NNNCFGISQVNLIVNEE580 pKa = 3.9IPILLNPEE588 pKa = 3.83EE589 pKa = 4.22DD590 pKa = 3.71VVYY593 pKa = 10.43CLNSFPEE600 pKa = 4.64TITLEE605 pKa = 4.13GNIFNNDD612 pKa = 3.28SEE614 pKa = 4.51NFSYY618 pKa = 10.33RR619 pKa = 11.84WSTGEE624 pKa = 3.94TTSSIEE630 pKa = 3.85VNQIGVFTLEE640 pKa = 3.93ITNSSGCSNVRR651 pKa = 11.84TITVLPSNVATIDD664 pKa = 3.76EE665 pKa = 4.88IIVNDD670 pKa = 3.63VTSSNTATINVTGEE684 pKa = 3.66GDD686 pKa = 3.63YY687 pKa = 11.47EE688 pKa = 4.09FALDD692 pKa = 3.71SLSGFFQDD700 pKa = 5.08EE701 pKa = 4.18NTFNNISPGFHH712 pKa = 5.28TVFVRR717 pKa = 11.84DD718 pKa = 3.61KK719 pKa = 9.59NGCGVVSKK727 pKa = 9.99LFSVIGFPRR736 pKa = 11.84FFTPNGDD743 pKa = 3.73SYY745 pKa = 11.97NDD747 pKa = 3.03TWQLSGVGNGFNEE760 pKa = 3.75NSLINIFDD768 pKa = 3.89RR769 pKa = 11.84FGKK772 pKa = 10.64LIIQIDD778 pKa = 3.76ANGRR782 pKa = 11.84GWDD785 pKa = 3.58GTFNGNDD792 pKa = 4.13LPASDD797 pKa = 3.34YY798 pKa = 9.47WFRR801 pKa = 11.84VRR803 pKa = 11.84LEE805 pKa = 4.03DD806 pKa = 3.47GRR808 pKa = 11.84IFSGHH813 pKa = 6.14FSLIRR818 pKa = 4.07
MM1 pKa = 7.68RR2 pKa = 11.84FISVFIFLIFFCFSNKK18 pKa = 7.88TFAQNIPPTILVEE31 pKa = 4.21NEE33 pKa = 3.72PFYY36 pKa = 11.47CPLSRR41 pKa = 11.84QNIVTAFDD49 pKa = 4.66IIDD52 pKa = 4.18PDD54 pKa = 3.79DD55 pKa = 4.12TEE57 pKa = 4.47VEE59 pKa = 4.03AFFIQISVGYY69 pKa = 10.41DD70 pKa = 2.9EE71 pKa = 5.98GQDD74 pKa = 3.72FLEE77 pKa = 4.43LTNSDD82 pKa = 3.88FHH84 pKa = 8.2PEE86 pKa = 4.01IVAIWNSEE94 pKa = 3.9EE95 pKa = 4.13GRR97 pKa = 11.84LTLEE101 pKa = 4.11GVGGIMIDD109 pKa = 3.45YY110 pKa = 9.78DD111 pKa = 4.39AIIAAVNDD119 pKa = 3.21IVFFNTDD126 pKa = 3.67PDD128 pKa = 3.75NTTSKK133 pKa = 10.56SFSYY137 pKa = 10.53TIGDD141 pKa = 3.93ANFLPSTGHH150 pKa = 6.87FYY152 pKa = 11.09QFIPDD157 pKa = 4.34IGISWIAAEE166 pKa = 4.08QAASNLTFFGLQGYY180 pKa = 8.27LATLTSAEE188 pKa = 4.13EE189 pKa = 3.79AQFAGNQITGNGWIGGSDD207 pKa = 3.25AAEE210 pKa = 3.89EE211 pKa = 4.69GVWRR215 pKa = 11.84WVTGPEE221 pKa = 3.55NGMVFWNGGVNGSTPNFANWNTGEE245 pKa = 4.01PNQLGNEE252 pKa = 4.3DD253 pKa = 3.98YY254 pKa = 11.79AHH256 pKa = 6.26ITSPGIGIDD265 pKa = 4.03GSWNDD270 pKa = 4.2LRR272 pKa = 11.84NEE274 pKa = 4.39GDD276 pKa = 3.18PSGPFQSMGYY286 pKa = 7.97VVEE289 pKa = 4.18YY290 pKa = 10.66GGLPGDD296 pKa = 3.55PVINISASTTFMIPTILSVSNDD318 pKa = 3.04INSCGPVEE326 pKa = 4.39VDD328 pKa = 3.97LTATINGEE336 pKa = 4.13RR337 pKa = 11.84IFWFASEE344 pKa = 4.1TSDD347 pKa = 4.59VILHH351 pKa = 5.97TGIPFTVNVEE361 pKa = 3.91NTTTFWVLASVDD373 pKa = 3.46GCLTGVRR380 pKa = 11.84TPVTVMIDD388 pKa = 3.53GLPSINNASLTQCDD402 pKa = 3.28VDD404 pKa = 4.24GVSDD408 pKa = 4.3GSTVFNLNEE417 pKa = 4.04ARR419 pKa = 11.84TALIGDD425 pKa = 4.0ITNVSIQFFTSLIDD439 pKa = 3.62AQNATNAVNSNVFRR453 pKa = 11.84NTVNPQTLFVRR464 pKa = 11.84ANDD467 pKa = 3.53NVSACFSIAEE477 pKa = 4.24LVLDD481 pKa = 4.42VSTTQVNNATLEE493 pKa = 4.19VCDD496 pKa = 3.94TDD498 pKa = 4.21GNEE501 pKa = 4.68DD502 pKa = 3.74GLHH505 pKa = 6.65LFTLSDD511 pKa = 4.06ADD513 pKa = 4.92DD514 pKa = 5.01EE515 pKa = 4.53ILNGLTNNGDD525 pKa = 3.48FDD527 pKa = 3.76IVYY530 pKa = 10.18YY531 pKa = 11.03ASTEE535 pKa = 4.1DD536 pKa = 5.22ALLEE540 pKa = 4.08QNPLNNDD547 pKa = 3.61YY548 pKa = 11.9ANVTPGNQIIYY559 pKa = 10.87ARR561 pKa = 11.84IEE563 pKa = 4.2DD564 pKa = 3.85NNNCFGISQVNLIVNEE580 pKa = 3.9IPILLNPEE588 pKa = 3.83EE589 pKa = 4.22DD590 pKa = 3.71VVYY593 pKa = 10.43CLNSFPEE600 pKa = 4.64TITLEE605 pKa = 4.13GNIFNNDD612 pKa = 3.28SEE614 pKa = 4.51NFSYY618 pKa = 10.33RR619 pKa = 11.84WSTGEE624 pKa = 3.94TTSSIEE630 pKa = 3.85VNQIGVFTLEE640 pKa = 3.93ITNSSGCSNVRR651 pKa = 11.84TITVLPSNVATIDD664 pKa = 3.76EE665 pKa = 4.88IIVNDD670 pKa = 3.63VTSSNTATINVTGEE684 pKa = 3.66GDD686 pKa = 3.63YY687 pKa = 11.47EE688 pKa = 4.09FALDD692 pKa = 3.71SLSGFFQDD700 pKa = 5.08EE701 pKa = 4.18NTFNNISPGFHH712 pKa = 5.28TVFVRR717 pKa = 11.84DD718 pKa = 3.61KK719 pKa = 9.59NGCGVVSKK727 pKa = 9.99LFSVIGFPRR736 pKa = 11.84FFTPNGDD743 pKa = 3.73SYY745 pKa = 11.97NDD747 pKa = 3.03TWQLSGVGNGFNEE760 pKa = 3.75NSLINIFDD768 pKa = 3.89RR769 pKa = 11.84FGKK772 pKa = 10.64LIIQIDD778 pKa = 3.76ANGRR782 pKa = 11.84GWDD785 pKa = 3.58GTFNGNDD792 pKa = 4.13LPASDD797 pKa = 3.34YY798 pKa = 9.47WFRR801 pKa = 11.84VRR803 pKa = 11.84LEE805 pKa = 4.03DD806 pKa = 3.47GRR808 pKa = 11.84IFSGHH813 pKa = 6.14FSLIRR818 pKa = 4.07
Molecular weight: 89.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6L9EWJ7|A0A6L9EWJ7_9FLAO Chorismate synthase OS=Flavobacteriaceae bacterium R38 OX=2697054 GN=aroC PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.52LSVSSEE47 pKa = 3.87PRR49 pKa = 11.84HH50 pKa = 5.92KK51 pKa = 10.61KK52 pKa = 9.84
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.52LSVSSEE47 pKa = 3.87PRR49 pKa = 11.84HH50 pKa = 5.92KK51 pKa = 10.61KK52 pKa = 9.84
Molecular weight: 6.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1175463 |
38 |
4194 |
332.0 |
37.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.001 ± 0.037 | 0.696 ± 0.011 |
5.603 ± 0.034 | 6.776 ± 0.043 |
5.382 ± 0.041 | 6.281 ± 0.044 |
1.761 ± 0.019 | 8.467 ± 0.042 |
8.022 ± 0.06 | 9.36 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.968 ± 0.021 | 6.335 ± 0.041 |
3.317 ± 0.027 | 3.239 ± 0.019 |
3.609 ± 0.029 | 6.472 ± 0.032 |
5.647 ± 0.055 | 6.003 ± 0.038 |
1.041 ± 0.018 | 4.022 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
