
Planctomycetes bacterium K22_7
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Pirellulaceae; Rubripirellula
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
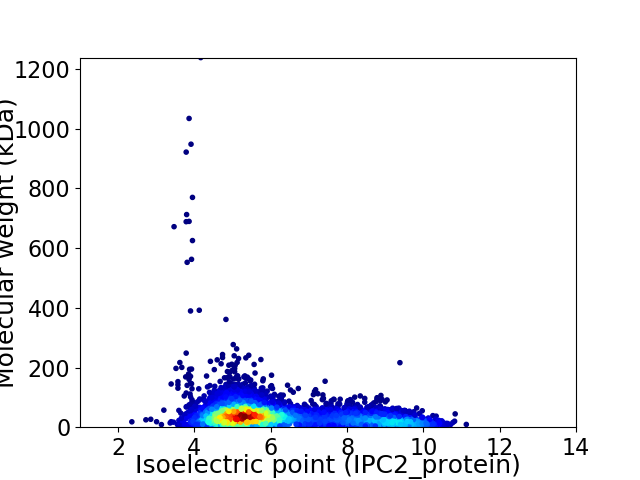
Virtual 2D-PAGE plot for 6373 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A517NAA3|A0A517NAA3_9BACT Uncharacterized protein OS=Planctomycetes bacterium K22_7 OX=1930273 GN=K227x_24480 PE=4 SV=1
MM1 pKa = 7.21KK2 pKa = 10.36RR3 pKa = 11.84STRR6 pKa = 11.84RR7 pKa = 11.84TKK9 pKa = 9.56NQRR12 pKa = 11.84TSLEE16 pKa = 4.05TLEE19 pKa = 5.46ARR21 pKa = 11.84DD22 pKa = 3.6LLAAFGTPWPEE33 pKa = 3.75ARR35 pKa = 11.84DD36 pKa = 3.63LSISFPADD44 pKa = 2.91EE45 pKa = 4.37VAIGEE50 pKa = 4.33YY51 pKa = 10.67EE52 pKa = 3.76NDD54 pKa = 2.79IRR56 pKa = 11.84QTLDD60 pKa = 4.06AADD63 pKa = 3.56QQQWQEE69 pKa = 3.82LALRR73 pKa = 11.84AYY75 pKa = 7.25QTWAIHH81 pKa = 6.41ADD83 pKa = 3.17INVGLRR89 pKa = 11.84NDD91 pKa = 3.5FDD93 pKa = 4.39VSFGAAGLMNQDD105 pKa = 2.86PRR107 pKa = 11.84FGEE110 pKa = 3.94FRR112 pKa = 11.84IGAFPQDD119 pKa = 2.93GLMANSLPFQVAAGTYY135 pKa = 10.38SGDD138 pKa = 3.59LLLNSNQDD146 pKa = 3.04YY147 pKa = 10.97RR148 pKa = 11.84LYY150 pKa = 10.87DD151 pKa = 3.39WPDD154 pKa = 3.01SVAPDD159 pKa = 4.1LSTWDD164 pKa = 3.19EE165 pKa = 4.06GQRR168 pKa = 11.84DD169 pKa = 4.16LFSVLLHH176 pKa = 6.14EE177 pKa = 5.52AGNTLGVDD185 pKa = 4.04DD186 pKa = 5.75NQMEE190 pKa = 4.32WSVMFGQYY198 pKa = 6.98TTPKK202 pKa = 10.19GVLAPEE208 pKa = 5.15DD209 pKa = 3.25IGEE212 pKa = 4.05IQQLYY217 pKa = 9.99GMRR220 pKa = 11.84SDD222 pKa = 4.08PYY224 pKa = 10.84EE225 pKa = 4.26SVTNDD230 pKa = 3.23QLATATIIPIPIGFAPASEE249 pKa = 4.63TIRR252 pKa = 11.84TRR254 pKa = 11.84GSIASANDD262 pKa = 2.76VDD264 pKa = 4.4HH265 pKa = 7.06FRR267 pKa = 11.84FSPVPGSNSVTIRR280 pKa = 11.84LNAEE284 pKa = 4.46GISLLEE290 pKa = 4.03SQIEE294 pKa = 4.18VTLADD299 pKa = 3.81GTVVAQSMAASVFEE313 pKa = 4.4NDD315 pKa = 3.52HH316 pKa = 5.27QVEE319 pKa = 4.3LTGLGVGSDD328 pKa = 3.79LFVKK332 pKa = 8.97VTAADD337 pKa = 3.58SDD339 pKa = 4.11SLYY342 pKa = 11.03SIGDD346 pKa = 3.58YY347 pKa = 10.88VLEE350 pKa = 4.32LDD352 pKa = 4.0YY353 pKa = 11.31RR354 pKa = 11.84DD355 pKa = 4.59AAVQSQDD362 pKa = 3.6IVPGDD367 pKa = 3.89YY368 pKa = 10.55QSGADD373 pKa = 3.58SLYY376 pKa = 11.11SGFDD380 pKa = 3.69LLDD383 pKa = 4.14LEE385 pKa = 4.97TDD387 pKa = 3.46VSTTASAIEE396 pKa = 4.08IDD398 pKa = 3.45AHH400 pKa = 6.91DD401 pKa = 4.98SIDD404 pKa = 3.57EE405 pKa = 4.07RR406 pKa = 11.84FEE408 pKa = 3.94IQSSVGSASDD418 pKa = 3.32VDD420 pKa = 3.69VWKK423 pKa = 10.93VVAPSTSQQRR433 pKa = 11.84LIVSLAGVGSGSPDD447 pKa = 3.32LDD449 pKa = 3.92LYY451 pKa = 11.62VVDD454 pKa = 5.57DD455 pKa = 3.82QGQSVGTTARR465 pKa = 11.84LRR467 pKa = 11.84PDD469 pKa = 3.39GTWTVEE475 pKa = 4.08VNQPTAGQEE484 pKa = 3.66YY485 pKa = 9.09FIRR488 pKa = 11.84VSVDD492 pKa = 2.71PGSAVEE498 pKa = 4.02VGNYY502 pKa = 7.96VAVAEE507 pKa = 4.74FVSPPAQMNDD517 pKa = 4.18LISGDD522 pKa = 3.13VDD524 pKa = 3.39ATVDD528 pKa = 3.66EE529 pKa = 4.94FFRR532 pKa = 11.84WTASKK537 pKa = 10.99SKK539 pKa = 10.55LFRR542 pKa = 11.84FDD544 pKa = 4.14FSAFGAAAGDD554 pKa = 4.0GARR557 pKa = 11.84LRR559 pKa = 11.84IYY561 pKa = 10.07DD562 pKa = 3.54AHH564 pKa = 4.84THH566 pKa = 6.3EE567 pKa = 4.44IRR569 pKa = 11.84LVIGAQNDD577 pKa = 3.46MTRR580 pKa = 11.84TGMAWLQQGDD590 pKa = 3.9YY591 pKa = 10.53VLRR594 pKa = 11.84FTAFSADD601 pKa = 3.92DD602 pKa = 3.54QSVAPIQFTLTCDD615 pKa = 4.86GISDD619 pKa = 4.07DD620 pKa = 4.67QDD622 pKa = 3.27EE623 pKa = 4.62TGYY626 pKa = 11.3DD627 pKa = 3.73PDD629 pKa = 4.71NDD631 pKa = 4.32PDD633 pKa = 4.08YY634 pKa = 11.59DD635 pKa = 4.14PYY637 pKa = 10.4TYY639 pKa = 10.32EE640 pKa = 4.03YY641 pKa = 9.5TYY643 pKa = 11.08HH644 pKa = 7.79DD645 pKa = 3.45YY646 pKa = 11.41GYY648 pKa = 10.44YY649 pKa = 8.71YY650 pKa = 11.13SNYY653 pKa = 9.22EE654 pKa = 3.89YY655 pKa = 11.06YY656 pKa = 10.8YY657 pKa = 11.01DD658 pKa = 3.2WDD660 pKa = 3.37YY661 pKa = 11.68YY662 pKa = 9.22YY663 pKa = 10.84QYY665 pKa = 11.45DD666 pKa = 3.66GDD668 pKa = 4.23PNYY671 pKa = 11.21AHH673 pKa = 7.04YY674 pKa = 11.19YY675 pKa = 7.98EE676 pKa = 4.61
MM1 pKa = 7.21KK2 pKa = 10.36RR3 pKa = 11.84STRR6 pKa = 11.84RR7 pKa = 11.84TKK9 pKa = 9.56NQRR12 pKa = 11.84TSLEE16 pKa = 4.05TLEE19 pKa = 5.46ARR21 pKa = 11.84DD22 pKa = 3.6LLAAFGTPWPEE33 pKa = 3.75ARR35 pKa = 11.84DD36 pKa = 3.63LSISFPADD44 pKa = 2.91EE45 pKa = 4.37VAIGEE50 pKa = 4.33YY51 pKa = 10.67EE52 pKa = 3.76NDD54 pKa = 2.79IRR56 pKa = 11.84QTLDD60 pKa = 4.06AADD63 pKa = 3.56QQQWQEE69 pKa = 3.82LALRR73 pKa = 11.84AYY75 pKa = 7.25QTWAIHH81 pKa = 6.41ADD83 pKa = 3.17INVGLRR89 pKa = 11.84NDD91 pKa = 3.5FDD93 pKa = 4.39VSFGAAGLMNQDD105 pKa = 2.86PRR107 pKa = 11.84FGEE110 pKa = 3.94FRR112 pKa = 11.84IGAFPQDD119 pKa = 2.93GLMANSLPFQVAAGTYY135 pKa = 10.38SGDD138 pKa = 3.59LLLNSNQDD146 pKa = 3.04YY147 pKa = 10.97RR148 pKa = 11.84LYY150 pKa = 10.87DD151 pKa = 3.39WPDD154 pKa = 3.01SVAPDD159 pKa = 4.1LSTWDD164 pKa = 3.19EE165 pKa = 4.06GQRR168 pKa = 11.84DD169 pKa = 4.16LFSVLLHH176 pKa = 6.14EE177 pKa = 5.52AGNTLGVDD185 pKa = 4.04DD186 pKa = 5.75NQMEE190 pKa = 4.32WSVMFGQYY198 pKa = 6.98TTPKK202 pKa = 10.19GVLAPEE208 pKa = 5.15DD209 pKa = 3.25IGEE212 pKa = 4.05IQQLYY217 pKa = 9.99GMRR220 pKa = 11.84SDD222 pKa = 4.08PYY224 pKa = 10.84EE225 pKa = 4.26SVTNDD230 pKa = 3.23QLATATIIPIPIGFAPASEE249 pKa = 4.63TIRR252 pKa = 11.84TRR254 pKa = 11.84GSIASANDD262 pKa = 2.76VDD264 pKa = 4.4HH265 pKa = 7.06FRR267 pKa = 11.84FSPVPGSNSVTIRR280 pKa = 11.84LNAEE284 pKa = 4.46GISLLEE290 pKa = 4.03SQIEE294 pKa = 4.18VTLADD299 pKa = 3.81GTVVAQSMAASVFEE313 pKa = 4.4NDD315 pKa = 3.52HH316 pKa = 5.27QVEE319 pKa = 4.3LTGLGVGSDD328 pKa = 3.79LFVKK332 pKa = 8.97VTAADD337 pKa = 3.58SDD339 pKa = 4.11SLYY342 pKa = 11.03SIGDD346 pKa = 3.58YY347 pKa = 10.88VLEE350 pKa = 4.32LDD352 pKa = 4.0YY353 pKa = 11.31RR354 pKa = 11.84DD355 pKa = 4.59AAVQSQDD362 pKa = 3.6IVPGDD367 pKa = 3.89YY368 pKa = 10.55QSGADD373 pKa = 3.58SLYY376 pKa = 11.11SGFDD380 pKa = 3.69LLDD383 pKa = 4.14LEE385 pKa = 4.97TDD387 pKa = 3.46VSTTASAIEE396 pKa = 4.08IDD398 pKa = 3.45AHH400 pKa = 6.91DD401 pKa = 4.98SIDD404 pKa = 3.57EE405 pKa = 4.07RR406 pKa = 11.84FEE408 pKa = 3.94IQSSVGSASDD418 pKa = 3.32VDD420 pKa = 3.69VWKK423 pKa = 10.93VVAPSTSQQRR433 pKa = 11.84LIVSLAGVGSGSPDD447 pKa = 3.32LDD449 pKa = 3.92LYY451 pKa = 11.62VVDD454 pKa = 5.57DD455 pKa = 3.82QGQSVGTTARR465 pKa = 11.84LRR467 pKa = 11.84PDD469 pKa = 3.39GTWTVEE475 pKa = 4.08VNQPTAGQEE484 pKa = 3.66YY485 pKa = 9.09FIRR488 pKa = 11.84VSVDD492 pKa = 2.71PGSAVEE498 pKa = 4.02VGNYY502 pKa = 7.96VAVAEE507 pKa = 4.74FVSPPAQMNDD517 pKa = 4.18LISGDD522 pKa = 3.13VDD524 pKa = 3.39ATVDD528 pKa = 3.66EE529 pKa = 4.94FFRR532 pKa = 11.84WTASKK537 pKa = 10.99SKK539 pKa = 10.55LFRR542 pKa = 11.84FDD544 pKa = 4.14FSAFGAAAGDD554 pKa = 4.0GARR557 pKa = 11.84LRR559 pKa = 11.84IYY561 pKa = 10.07DD562 pKa = 3.54AHH564 pKa = 4.84THH566 pKa = 6.3EE567 pKa = 4.44IRR569 pKa = 11.84LVIGAQNDD577 pKa = 3.46MTRR580 pKa = 11.84TGMAWLQQGDD590 pKa = 3.9YY591 pKa = 10.53VLRR594 pKa = 11.84FTAFSADD601 pKa = 3.92DD602 pKa = 3.54QSVAPIQFTLTCDD615 pKa = 4.86GISDD619 pKa = 4.07DD620 pKa = 4.67QDD622 pKa = 3.27EE623 pKa = 4.62TGYY626 pKa = 11.3DD627 pKa = 3.73PDD629 pKa = 4.71NDD631 pKa = 4.32PDD633 pKa = 4.08YY634 pKa = 11.59DD635 pKa = 4.14PYY637 pKa = 10.4TYY639 pKa = 10.32EE640 pKa = 4.03YY641 pKa = 9.5TYY643 pKa = 11.08HH644 pKa = 7.79DD645 pKa = 3.45YY646 pKa = 11.41GYY648 pKa = 10.44YY649 pKa = 8.71YY650 pKa = 11.13SNYY653 pKa = 9.22EE654 pKa = 3.89YY655 pKa = 11.06YY656 pKa = 10.8YY657 pKa = 11.01DD658 pKa = 3.2WDD660 pKa = 3.37YY661 pKa = 11.68YY662 pKa = 9.22YY663 pKa = 10.84QYY665 pKa = 11.45DD666 pKa = 3.66GDD668 pKa = 4.23PNYY671 pKa = 11.21AHH673 pKa = 7.04YY674 pKa = 11.19YY675 pKa = 7.98EE676 pKa = 4.61
Molecular weight: 74.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A517NIX7|A0A517NIX7_9BACT Copper-sensing transcriptional repressor CsoR OS=Planctomycetes bacterium K22_7 OX=1930273 GN=csoR_2 PE=4 SV=1
MM1 pKa = 7.41GMTVLSGLGQSAIRR15 pKa = 11.84SVMGMTVHH23 pKa = 6.78SGLGQSTIRR32 pKa = 11.84SVMGMTVHH40 pKa = 6.9SGLGQSAIRR49 pKa = 11.84SVMGMTGLSALGQSAIRR66 pKa = 11.84LVMGMTVHH74 pKa = 6.71SGLGQSTIRR83 pKa = 11.84LVMGMTVHH91 pKa = 6.71SGLGQSTIRR100 pKa = 11.84SVMGMTVHH108 pKa = 6.9SGLGQSAIRR117 pKa = 11.84SVMGMTVHH125 pKa = 6.78SGLGQSTIRR134 pKa = 11.84SVMGMTVHH142 pKa = 6.9SGLGQSAIRR151 pKa = 11.84SVMGMTVHH159 pKa = 6.78SGLGQSTIRR168 pKa = 11.84SVMGMTVHH176 pKa = 6.9SGLGQSAIRR185 pKa = 11.84SVMGMTVHH193 pKa = 6.9SGLGQSAIRR202 pKa = 11.84LVMGMTVLSGLGQSAIRR219 pKa = 11.84SVMGMTVHH227 pKa = 6.9SGLGQSAIRR236 pKa = 11.84LVMGMTVHH244 pKa = 6.83SGLGQSAIRR253 pKa = 11.84SVMGMTVHH261 pKa = 6.78SGLGQSTIRR270 pKa = 11.84SVMGMTVHH278 pKa = 6.9SGLGQSAIRR287 pKa = 11.84SVMGMTVHH295 pKa = 6.78SGLGQSTIRR304 pKa = 11.84SVMGMTVHH312 pKa = 6.78SGLGQSTIRR321 pKa = 11.84SVMGMTGLSALGQSAIRR338 pKa = 11.84SVMGMTVLSVLGQSTIRR355 pKa = 11.84LVVGMTVLSVLGQSTIRR372 pKa = 11.84LVVRR376 pKa = 11.84FATRR380 pKa = 11.84LAGSQDD386 pKa = 3.42VQRR389 pKa = 11.84RR390 pKa = 11.84AASCVCKK397 pKa = 10.73SKK399 pKa = 10.69INCPSSVMAAVSRR412 pKa = 11.84INAASLVGTWRR423 pKa = 11.84ILCAASRR430 pKa = 11.84AVGIQRR436 pKa = 3.87
MM1 pKa = 7.41GMTVLSGLGQSAIRR15 pKa = 11.84SVMGMTVHH23 pKa = 6.78SGLGQSTIRR32 pKa = 11.84SVMGMTVHH40 pKa = 6.9SGLGQSAIRR49 pKa = 11.84SVMGMTGLSALGQSAIRR66 pKa = 11.84LVMGMTVHH74 pKa = 6.71SGLGQSTIRR83 pKa = 11.84LVMGMTVHH91 pKa = 6.71SGLGQSTIRR100 pKa = 11.84SVMGMTVHH108 pKa = 6.9SGLGQSAIRR117 pKa = 11.84SVMGMTVHH125 pKa = 6.78SGLGQSTIRR134 pKa = 11.84SVMGMTVHH142 pKa = 6.9SGLGQSAIRR151 pKa = 11.84SVMGMTVHH159 pKa = 6.78SGLGQSTIRR168 pKa = 11.84SVMGMTVHH176 pKa = 6.9SGLGQSAIRR185 pKa = 11.84SVMGMTVHH193 pKa = 6.9SGLGQSAIRR202 pKa = 11.84LVMGMTVLSGLGQSAIRR219 pKa = 11.84SVMGMTVHH227 pKa = 6.9SGLGQSAIRR236 pKa = 11.84LVMGMTVHH244 pKa = 6.83SGLGQSAIRR253 pKa = 11.84SVMGMTVHH261 pKa = 6.78SGLGQSTIRR270 pKa = 11.84SVMGMTVHH278 pKa = 6.9SGLGQSAIRR287 pKa = 11.84SVMGMTVHH295 pKa = 6.78SGLGQSTIRR304 pKa = 11.84SVMGMTVHH312 pKa = 6.78SGLGQSTIRR321 pKa = 11.84SVMGMTGLSALGQSAIRR338 pKa = 11.84SVMGMTVLSVLGQSTIRR355 pKa = 11.84LVVGMTVLSVLGQSTIRR372 pKa = 11.84LVVRR376 pKa = 11.84FATRR380 pKa = 11.84LAGSQDD386 pKa = 3.42VQRR389 pKa = 11.84RR390 pKa = 11.84AASCVCKK397 pKa = 10.73SKK399 pKa = 10.69INCPSSVMAAVSRR412 pKa = 11.84INAASLVGTWRR423 pKa = 11.84ILCAASRR430 pKa = 11.84AVGIQRR436 pKa = 3.87
Molecular weight: 44.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2431915 |
29 |
11647 |
381.6 |
41.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.91 ± 0.034 | 1.094 ± 0.015 |
6.858 ± 0.039 | 5.305 ± 0.027 |
3.597 ± 0.017 | 7.896 ± 0.044 |
2.19 ± 0.019 | 5.214 ± 0.023 |
3.304 ± 0.027 | 9.099 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.375 ± 0.02 | 3.155 ± 0.027 |
5.285 ± 0.028 | 4.189 ± 0.022 |
6.652 ± 0.042 | 7.058 ± 0.029 |
5.797 ± 0.044 | 7.268 ± 0.029 |
1.494 ± 0.017 | 2.259 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
