
Corynebacterium testudinoris
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Corynebacteriaceae;
Average proteome isoelectric point is 5.84
Get precalculated fractions of proteins
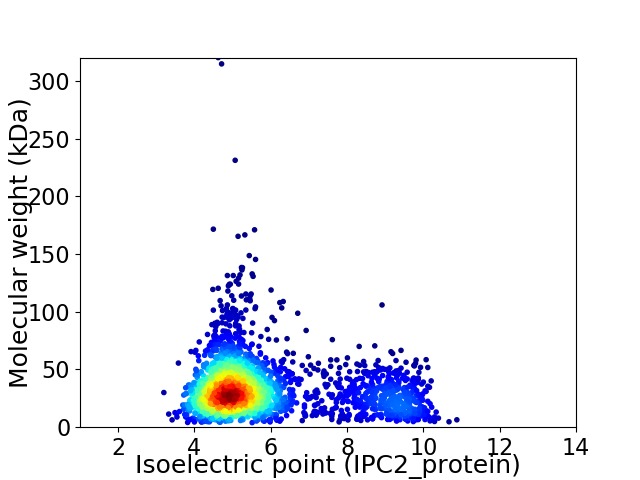
Virtual 2D-PAGE plot for 2543 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
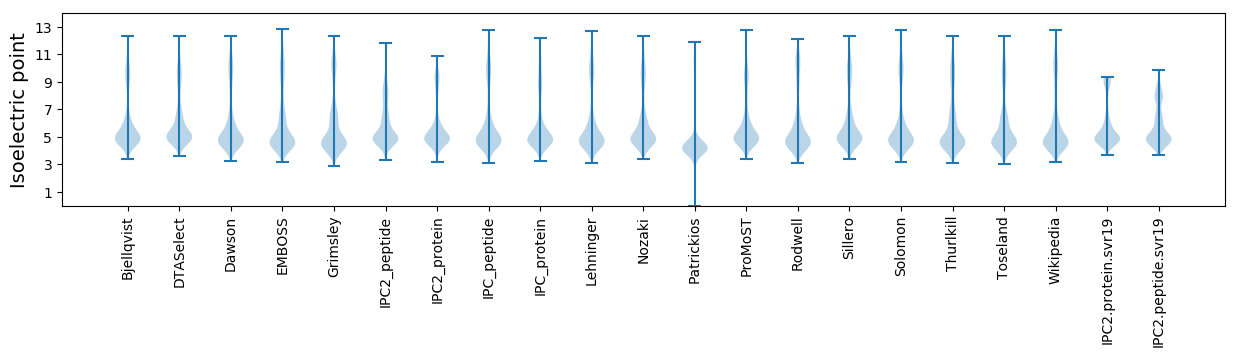
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G3HF05|A0A0G3HF05_9CORY DNA-binding protein excisionase family OS=Corynebacterium testudinoris OX=136857 GN=CTEST_11500 PE=4 SV=1
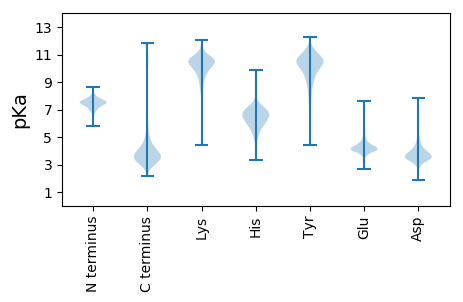
MM1 pKa = 7.58ARR3 pKa = 11.84TGSPIWIDD11 pKa = 4.16LGTHH15 pKa = 7.16DD16 pKa = 4.78IDD18 pKa = 4.77GSVSFYY24 pKa = 10.96SQVFGWDD31 pKa = 3.47VPEE34 pKa = 4.6GAAEE38 pKa = 3.94FGGYY42 pKa = 10.15RR43 pKa = 11.84IATKK47 pKa = 10.64NGVPVGGMMSSLMGPDD63 pKa = 3.83GPLEE67 pKa = 4.13EE68 pKa = 5.07PEE70 pKa = 4.98YY71 pKa = 8.26PTAWTVYY78 pKa = 10.35LAVEE82 pKa = 4.32DD83 pKa = 4.19CAAAVAAAEE92 pKa = 4.33KK93 pKa = 10.62AGATVVVPAMEE104 pKa = 4.2VGEE107 pKa = 4.55LGSMAILFDD116 pKa = 3.85PAGAGVGFWEE126 pKa = 4.43AKK128 pKa = 9.43EE129 pKa = 4.07FPGLAFNGQPGTPVWFEE146 pKa = 3.75QMSKK150 pKa = 10.86DD151 pKa = 3.6FQAAADD157 pKa = 5.01FYY159 pKa = 11.54QEE161 pKa = 3.85ALGWNLAYY169 pKa = 9.98MGEE172 pKa = 4.5DD173 pKa = 3.62GQPVDD178 pKa = 4.52SPPTSGIRR186 pKa = 11.84YY187 pKa = 5.9ATNGAGEE194 pKa = 4.16EE195 pKa = 4.16ASAGLCEE202 pKa = 4.87ADD204 pKa = 3.18SFLPAEE210 pKa = 4.47VPSYY214 pKa = 7.79WRR216 pKa = 11.84AYY218 pKa = 9.72IGVSDD223 pKa = 4.35TDD225 pKa = 3.14ATAAQIVEE233 pKa = 4.3LGGAVLDD240 pKa = 4.8GPMDD244 pKa = 3.86SPFGRR249 pKa = 11.84VATVADD255 pKa = 3.97PQGGTFQIVSVEE267 pKa = 4.05GG268 pKa = 3.38
MM1 pKa = 7.58ARR3 pKa = 11.84TGSPIWIDD11 pKa = 4.16LGTHH15 pKa = 7.16DD16 pKa = 4.78IDD18 pKa = 4.77GSVSFYY24 pKa = 10.96SQVFGWDD31 pKa = 3.47VPEE34 pKa = 4.6GAAEE38 pKa = 3.94FGGYY42 pKa = 10.15RR43 pKa = 11.84IATKK47 pKa = 10.64NGVPVGGMMSSLMGPDD63 pKa = 3.83GPLEE67 pKa = 4.13EE68 pKa = 5.07PEE70 pKa = 4.98YY71 pKa = 8.26PTAWTVYY78 pKa = 10.35LAVEE82 pKa = 4.32DD83 pKa = 4.19CAAAVAAAEE92 pKa = 4.33KK93 pKa = 10.62AGATVVVPAMEE104 pKa = 4.2VGEE107 pKa = 4.55LGSMAILFDD116 pKa = 3.85PAGAGVGFWEE126 pKa = 4.43AKK128 pKa = 9.43EE129 pKa = 4.07FPGLAFNGQPGTPVWFEE146 pKa = 3.75QMSKK150 pKa = 10.86DD151 pKa = 3.6FQAAADD157 pKa = 5.01FYY159 pKa = 11.54QEE161 pKa = 3.85ALGWNLAYY169 pKa = 9.98MGEE172 pKa = 4.5DD173 pKa = 3.62GQPVDD178 pKa = 4.52SPPTSGIRR186 pKa = 11.84YY187 pKa = 5.9ATNGAGEE194 pKa = 4.16EE195 pKa = 4.16ASAGLCEE202 pKa = 4.87ADD204 pKa = 3.18SFLPAEE210 pKa = 4.47VPSYY214 pKa = 7.79WRR216 pKa = 11.84AYY218 pKa = 9.72IGVSDD223 pKa = 4.35TDD225 pKa = 3.14ATAAQIVEE233 pKa = 4.3LGGAVLDD240 pKa = 4.8GPMDD244 pKa = 3.86SPFGRR249 pKa = 11.84VATVADD255 pKa = 3.97PQGGTFQIVSVEE267 pKa = 4.05GG268 pKa = 3.38
Molecular weight: 27.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G3H5Q1|A0A0G3H5Q1_9CORY Sphingosine/diacylglycerol kinase-like enzyme OS=Corynebacterium testudinoris OX=136857 GN=CTEST_06335 PE=4 SV=1
MM1 pKa = 7.41TISPARR7 pKa = 11.84LIALTLTWATIWSGAFVAMKK27 pKa = 10.16VASQSAGAFAIVGIRR42 pKa = 11.84CTIAGAIMIAAAWGARR58 pKa = 11.84KK59 pKa = 9.64ALTRR63 pKa = 11.84SGVKK67 pKa = 8.61GTIVIGILNNLGYY80 pKa = 10.48LGLMASAMPYY90 pKa = 10.1VSAGMAAIITALTPLAVLVISALRR114 pKa = 11.84GRR116 pKa = 11.84ALLITQWIGCVVGFLGVVGSAWTRR140 pKa = 11.84LDD142 pKa = 3.34SGEE145 pKa = 4.24TQWRR149 pKa = 11.84GILFALGAVACLVAGTVLTPKK170 pKa = 10.06LVPPGSPWLSTGIQSLAAGIPALGLSLLFEE200 pKa = 4.57DD201 pKa = 5.01TPTFSTPFIVAEE213 pKa = 4.46AYY215 pKa = 10.53LILGASVLGMTMWLQLIRR233 pKa = 11.84EE234 pKa = 4.74AGPDD238 pKa = 3.25RR239 pKa = 11.84AAVAHH244 pKa = 6.6FLPPLISIALGALILGEE261 pKa = 4.24AVSPLALAMCIPVAIGVALATRR283 pKa = 11.84RR284 pKa = 11.84PRR286 pKa = 11.84KK287 pKa = 9.21PRR289 pKa = 11.84QLL291 pKa = 3.42
MM1 pKa = 7.41TISPARR7 pKa = 11.84LIALTLTWATIWSGAFVAMKK27 pKa = 10.16VASQSAGAFAIVGIRR42 pKa = 11.84CTIAGAIMIAAAWGARR58 pKa = 11.84KK59 pKa = 9.64ALTRR63 pKa = 11.84SGVKK67 pKa = 8.61GTIVIGILNNLGYY80 pKa = 10.48LGLMASAMPYY90 pKa = 10.1VSAGMAAIITALTPLAVLVISALRR114 pKa = 11.84GRR116 pKa = 11.84ALLITQWIGCVVGFLGVVGSAWTRR140 pKa = 11.84LDD142 pKa = 3.34SGEE145 pKa = 4.24TQWRR149 pKa = 11.84GILFALGAVACLVAGTVLTPKK170 pKa = 10.06LVPPGSPWLSTGIQSLAAGIPALGLSLLFEE200 pKa = 4.57DD201 pKa = 5.01TPTFSTPFIVAEE213 pKa = 4.46AYY215 pKa = 10.53LILGASVLGMTMWLQLIRR233 pKa = 11.84EE234 pKa = 4.74AGPDD238 pKa = 3.25RR239 pKa = 11.84AAVAHH244 pKa = 6.6FLPPLISIALGALILGEE261 pKa = 4.24AVSPLALAMCIPVAIGVALATRR283 pKa = 11.84RR284 pKa = 11.84PRR286 pKa = 11.84KK287 pKa = 9.21PRR289 pKa = 11.84QLL291 pKa = 3.42
Molecular weight: 29.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
818509 |
39 |
3015 |
321.9 |
34.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.736 ± 0.056 | 0.629 ± 0.013 |
6.15 ± 0.042 | 6.004 ± 0.053 |
3.304 ± 0.03 | 8.489 ± 0.044 |
2.157 ± 0.024 | 5.175 ± 0.035 |
2.576 ± 0.033 | 10.108 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.227 ± 0.018 | 2.598 ± 0.027 |
5.193 ± 0.03 | 3.114 ± 0.03 |
6.444 ± 0.041 | 5.886 ± 0.031 |
6.192 ± 0.034 | 8.417 ± 0.05 |
1.462 ± 0.021 | 2.138 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
