
Ensifer adhaerens OV14
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Sinorhizobium/Ensifer group; Ensifer; Ensifer adhaerens
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
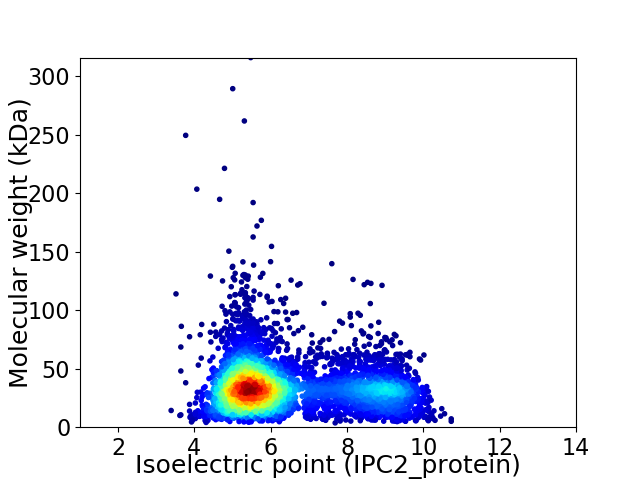
Virtual 2D-PAGE plot for 5422 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W8IPW3|W8IPW3_ENSAD HTH gntR-type domain-containing protein OS=Ensifer adhaerens OV14 OX=1416753 GN=OV14_b0279 PE=3 SV=1
MM1 pKa = 7.99LYY3 pKa = 8.87GTEE6 pKa = 4.45ANDD9 pKa = 4.61SIWGDD14 pKa = 3.42GSVDD18 pKa = 3.18VTMAGGKK25 pKa = 10.29GDD27 pKa = 4.93DD28 pKa = 3.25IYY30 pKa = 11.78YY31 pKa = 10.19LYY33 pKa = 11.08SGINRR38 pKa = 11.84AMEE41 pKa = 3.91TAGAGVDD48 pKa = 4.64TINTWMSYY56 pKa = 7.27TLPEE60 pKa = 4.06NFEE63 pKa = 4.34NLTVTGSGRR72 pKa = 11.84HH73 pKa = 5.03AFGNNADD80 pKa = 4.45NIISGASGSQTIDD93 pKa = 2.86GGGGNDD99 pKa = 3.11ILVGGSGADD108 pKa = 2.9IFVVGRR114 pKa = 11.84GGGSDD119 pKa = 4.94LIADD123 pKa = 4.71FDD125 pKa = 4.41SSDD128 pKa = 3.45TVRR131 pKa = 11.84LNGYY135 pKa = 9.79DD136 pKa = 3.36LTSFQQVTSLATQEE150 pKa = 4.24GTDD153 pKa = 3.64LRR155 pKa = 11.84LNLGDD160 pKa = 4.18GEE162 pKa = 4.68SLLFADD168 pKa = 4.16TTVADD173 pKa = 4.65LAPEE177 pKa = 3.99QFQLSLDD184 pKa = 3.74RR185 pKa = 11.84SNLALSFADD194 pKa = 3.97EE195 pKa = 4.33FDD197 pKa = 3.92SLSLHH202 pKa = 7.41DD203 pKa = 4.89GDD205 pKa = 5.12QGVWDD210 pKa = 4.54AKK212 pKa = 10.4FWWAPEE218 pKa = 3.94KK219 pKa = 11.08GGTLASNGEE228 pKa = 4.32LQWYY232 pKa = 9.0INPDD236 pKa = 3.44HH237 pKa = 7.31AEE239 pKa = 3.85TAAINPFSVEE249 pKa = 4.36DD250 pKa = 3.83GVLTIKK256 pKa = 10.96AEE258 pKa = 4.05PTPDD262 pKa = 3.37ALKK265 pKa = 10.78PEE267 pKa = 4.18IDD269 pKa = 4.0GYY271 pKa = 11.3DD272 pKa = 3.61YY273 pKa = 11.21TSGMLTTYY281 pKa = 9.61STFAQTYY288 pKa = 8.16GYY290 pKa = 10.67FEE292 pKa = 4.65IRR294 pKa = 11.84ADD296 pKa = 3.66MPTEE300 pKa = 4.11TGTWPAFWLLPADD313 pKa = 4.39GSWPPEE319 pKa = 3.76LDD321 pKa = 3.52VVEE324 pKa = 4.8MRR326 pKa = 11.84GQQPNIVNVTVHH338 pKa = 6.09SNEE341 pKa = 4.11TGEE344 pKa = 4.3QTSEE348 pKa = 3.77TTAVKK353 pKa = 10.24VASTEE358 pKa = 4.03GFHH361 pKa = 6.57TYY363 pKa = 9.81GVLWEE368 pKa = 4.06EE369 pKa = 5.38DD370 pKa = 4.04EE371 pKa = 4.46IVWYY375 pKa = 10.52YY376 pKa = 11.57DD377 pKa = 3.24DD378 pKa = 4.05VAVARR383 pKa = 11.84ADD385 pKa = 3.96TPADD389 pKa = 3.41MHH391 pKa = 7.28DD392 pKa = 3.64PMYY395 pKa = 10.32MVVNLAVGGAAGEE408 pKa = 4.2PNADD412 pKa = 3.67FGDD415 pKa = 4.16GAEE418 pKa = 4.03MQIDD422 pKa = 4.51YY423 pKa = 10.7IHH425 pKa = 7.39AYY427 pKa = 10.27ALDD430 pKa = 4.16EE431 pKa = 4.37VQSATGEE438 pKa = 4.09AQPMDD443 pKa = 3.68DD444 pKa = 3.68WLSS447 pKa = 3.34
MM1 pKa = 7.99LYY3 pKa = 8.87GTEE6 pKa = 4.45ANDD9 pKa = 4.61SIWGDD14 pKa = 3.42GSVDD18 pKa = 3.18VTMAGGKK25 pKa = 10.29GDD27 pKa = 4.93DD28 pKa = 3.25IYY30 pKa = 11.78YY31 pKa = 10.19LYY33 pKa = 11.08SGINRR38 pKa = 11.84AMEE41 pKa = 3.91TAGAGVDD48 pKa = 4.64TINTWMSYY56 pKa = 7.27TLPEE60 pKa = 4.06NFEE63 pKa = 4.34NLTVTGSGRR72 pKa = 11.84HH73 pKa = 5.03AFGNNADD80 pKa = 4.45NIISGASGSQTIDD93 pKa = 2.86GGGGNDD99 pKa = 3.11ILVGGSGADD108 pKa = 2.9IFVVGRR114 pKa = 11.84GGGSDD119 pKa = 4.94LIADD123 pKa = 4.71FDD125 pKa = 4.41SSDD128 pKa = 3.45TVRR131 pKa = 11.84LNGYY135 pKa = 9.79DD136 pKa = 3.36LTSFQQVTSLATQEE150 pKa = 4.24GTDD153 pKa = 3.64LRR155 pKa = 11.84LNLGDD160 pKa = 4.18GEE162 pKa = 4.68SLLFADD168 pKa = 4.16TTVADD173 pKa = 4.65LAPEE177 pKa = 3.99QFQLSLDD184 pKa = 3.74RR185 pKa = 11.84SNLALSFADD194 pKa = 3.97EE195 pKa = 4.33FDD197 pKa = 3.92SLSLHH202 pKa = 7.41DD203 pKa = 4.89GDD205 pKa = 5.12QGVWDD210 pKa = 4.54AKK212 pKa = 10.4FWWAPEE218 pKa = 3.94KK219 pKa = 11.08GGTLASNGEE228 pKa = 4.32LQWYY232 pKa = 9.0INPDD236 pKa = 3.44HH237 pKa = 7.31AEE239 pKa = 3.85TAAINPFSVEE249 pKa = 4.36DD250 pKa = 3.83GVLTIKK256 pKa = 10.96AEE258 pKa = 4.05PTPDD262 pKa = 3.37ALKK265 pKa = 10.78PEE267 pKa = 4.18IDD269 pKa = 4.0GYY271 pKa = 11.3DD272 pKa = 3.61YY273 pKa = 11.21TSGMLTTYY281 pKa = 9.61STFAQTYY288 pKa = 8.16GYY290 pKa = 10.67FEE292 pKa = 4.65IRR294 pKa = 11.84ADD296 pKa = 3.66MPTEE300 pKa = 4.11TGTWPAFWLLPADD313 pKa = 4.39GSWPPEE319 pKa = 3.76LDD321 pKa = 3.52VVEE324 pKa = 4.8MRR326 pKa = 11.84GQQPNIVNVTVHH338 pKa = 6.09SNEE341 pKa = 4.11TGEE344 pKa = 4.3QTSEE348 pKa = 3.77TTAVKK353 pKa = 10.24VASTEE358 pKa = 4.03GFHH361 pKa = 6.57TYY363 pKa = 9.81GVLWEE368 pKa = 4.06EE369 pKa = 5.38DD370 pKa = 4.04EE371 pKa = 4.46IVWYY375 pKa = 10.52YY376 pKa = 11.57DD377 pKa = 3.24DD378 pKa = 4.05VAVARR383 pKa = 11.84ADD385 pKa = 3.96TPADD389 pKa = 3.41MHH391 pKa = 7.28DD392 pKa = 3.64PMYY395 pKa = 10.32MVVNLAVGGAAGEE408 pKa = 4.2PNADD412 pKa = 3.67FGDD415 pKa = 4.16GAEE418 pKa = 4.03MQIDD422 pKa = 4.51YY423 pKa = 10.7IHH425 pKa = 7.39AYY427 pKa = 10.27ALDD430 pKa = 4.16EE431 pKa = 4.37VQSATGEE438 pKa = 4.09AQPMDD443 pKa = 3.68DD444 pKa = 3.68WLSS447 pKa = 3.34
Molecular weight: 48.05 kDa
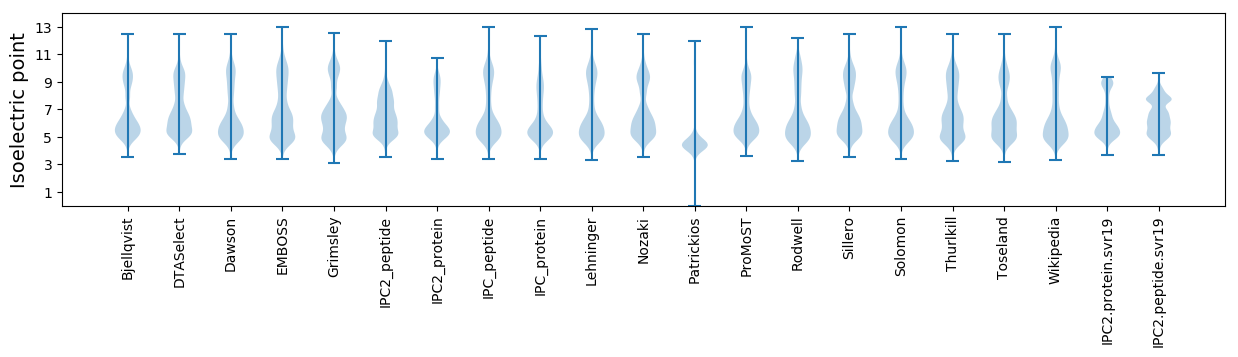
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8IB21|W8IB21_ENSAD Dipeptide ABC transport system permease protein DppB OS=Ensifer adhaerens OV14 OX=1416753 GN=dppB PE=3 SV=1
MM1 pKa = 7.55LLLHH5 pKa = 6.72KK6 pKa = 10.74SSFLVGALCFLYY18 pKa = 10.67LRR20 pKa = 11.84WQSPRR25 pKa = 11.84NEE27 pKa = 4.17GLGILAAGFLFLALASTLSGYY48 pKa = 10.46GEE50 pKa = 4.31QKK52 pKa = 10.68LLPAILWTLSSFTLGLGGYY71 pKa = 9.48ALFWIGLRR79 pKa = 11.84RR80 pKa = 11.84LSGQRR85 pKa = 11.84RR86 pKa = 11.84HH87 pKa = 5.83RR88 pKa = 11.84ADD90 pKa = 3.16WLALAIPLTGAIAGLITHH108 pKa = 6.32FHH110 pKa = 6.55VINSIRR116 pKa = 11.84ASAFNTGAFLFLAASAFAVVADD138 pKa = 5.18RR139 pKa = 11.84KK140 pKa = 10.42SEE142 pKa = 4.02PLPVRR147 pKa = 11.84TIFAASLAIGATLCLAVVVGLLKK170 pKa = 10.37PDD172 pKa = 3.66ILPITTTYY180 pKa = 11.59AFFLLIISHH189 pKa = 6.44FAIALFALVLVKK201 pKa = 10.33EE202 pKa = 4.22RR203 pKa = 11.84AEE205 pKa = 3.82AALRR209 pKa = 11.84RR210 pKa = 11.84TADD213 pKa = 3.07TDD215 pKa = 3.39MLTGVGNRR223 pKa = 11.84RR224 pKa = 11.84WFLSQLPRR232 pKa = 11.84FGWKK236 pKa = 9.85GDD238 pKa = 3.44ALAVMDD244 pKa = 5.16LDD246 pKa = 3.72FFKK249 pKa = 10.82RR250 pKa = 11.84INDD253 pKa = 3.93RR254 pKa = 11.84YY255 pKa = 9.3GHH257 pKa = 5.26QAGDD261 pKa = 3.37QVLIAFAEE269 pKa = 4.24LVRR272 pKa = 11.84ANLRR276 pKa = 11.84ATDD279 pKa = 3.85CFARR283 pKa = 11.84LGGVRR288 pKa = 11.84ALPSEE293 pKa = 4.21NEE295 pKa = 3.93RR296 pKa = 11.84GAGASDD302 pKa = 3.16RR303 pKa = 11.84RR304 pKa = 11.84AVAACHH310 pKa = 5.26RR311 pKa = 11.84TIAHH315 pKa = 6.79PLRR318 pKa = 11.84RR319 pKa = 11.84PAHH322 pKa = 6.15CRR324 pKa = 11.84QCQHH328 pKa = 5.94RR329 pKa = 11.84VCRR332 pKa = 11.84RR333 pKa = 11.84TGCEE337 pKa = 3.74KK338 pKa = 10.52QLGRR342 pKa = 11.84ALQYY346 pKa = 10.49GRR348 pKa = 11.84CRR350 pKa = 11.84TLSGQGCWPP359 pKa = 3.63
MM1 pKa = 7.55LLLHH5 pKa = 6.72KK6 pKa = 10.74SSFLVGALCFLYY18 pKa = 10.67LRR20 pKa = 11.84WQSPRR25 pKa = 11.84NEE27 pKa = 4.17GLGILAAGFLFLALASTLSGYY48 pKa = 10.46GEE50 pKa = 4.31QKK52 pKa = 10.68LLPAILWTLSSFTLGLGGYY71 pKa = 9.48ALFWIGLRR79 pKa = 11.84RR80 pKa = 11.84LSGQRR85 pKa = 11.84RR86 pKa = 11.84HH87 pKa = 5.83RR88 pKa = 11.84ADD90 pKa = 3.16WLALAIPLTGAIAGLITHH108 pKa = 6.32FHH110 pKa = 6.55VINSIRR116 pKa = 11.84ASAFNTGAFLFLAASAFAVVADD138 pKa = 5.18RR139 pKa = 11.84KK140 pKa = 10.42SEE142 pKa = 4.02PLPVRR147 pKa = 11.84TIFAASLAIGATLCLAVVVGLLKK170 pKa = 10.37PDD172 pKa = 3.66ILPITTTYY180 pKa = 11.59AFFLLIISHH189 pKa = 6.44FAIALFALVLVKK201 pKa = 10.33EE202 pKa = 4.22RR203 pKa = 11.84AEE205 pKa = 3.82AALRR209 pKa = 11.84RR210 pKa = 11.84TADD213 pKa = 3.07TDD215 pKa = 3.39MLTGVGNRR223 pKa = 11.84RR224 pKa = 11.84WFLSQLPRR232 pKa = 11.84FGWKK236 pKa = 9.85GDD238 pKa = 3.44ALAVMDD244 pKa = 5.16LDD246 pKa = 3.72FFKK249 pKa = 10.82RR250 pKa = 11.84INDD253 pKa = 3.93RR254 pKa = 11.84YY255 pKa = 9.3GHH257 pKa = 5.26QAGDD261 pKa = 3.37QVLIAFAEE269 pKa = 4.24LVRR272 pKa = 11.84ANLRR276 pKa = 11.84ATDD279 pKa = 3.85CFARR283 pKa = 11.84LGGVRR288 pKa = 11.84ALPSEE293 pKa = 4.21NEE295 pKa = 3.93RR296 pKa = 11.84GAGASDD302 pKa = 3.16RR303 pKa = 11.84RR304 pKa = 11.84AVAACHH310 pKa = 5.26RR311 pKa = 11.84TIAHH315 pKa = 6.79PLRR318 pKa = 11.84RR319 pKa = 11.84PAHH322 pKa = 6.15CRR324 pKa = 11.84QCQHH328 pKa = 5.94RR329 pKa = 11.84VCRR332 pKa = 11.84RR333 pKa = 11.84TGCEE337 pKa = 3.74KK338 pKa = 10.52QLGRR342 pKa = 11.84ALQYY346 pKa = 10.49GRR348 pKa = 11.84CRR350 pKa = 11.84TLSGQGCWPP359 pKa = 3.63
Molecular weight: 39.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1788006 |
34 |
2833 |
329.8 |
35.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.942 ± 0.041 | 0.793 ± 0.009 |
5.665 ± 0.026 | 5.76 ± 0.028 |
3.958 ± 0.023 | 8.418 ± 0.029 |
1.993 ± 0.018 | 5.699 ± 0.025 |
3.61 ± 0.028 | 10.08 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.605 ± 0.015 | 2.803 ± 0.017 |
4.797 ± 0.021 | 3.044 ± 0.018 |
6.715 ± 0.031 | 5.617 ± 0.023 |
5.384 ± 0.021 | 7.54 ± 0.025 |
1.25 ± 0.012 | 2.305 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
