
Planctomycetaceae bacterium SCGC AG-212-F19
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Planctomycetales; Planctomycetaceae; unclassified Planctomycetaceae
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
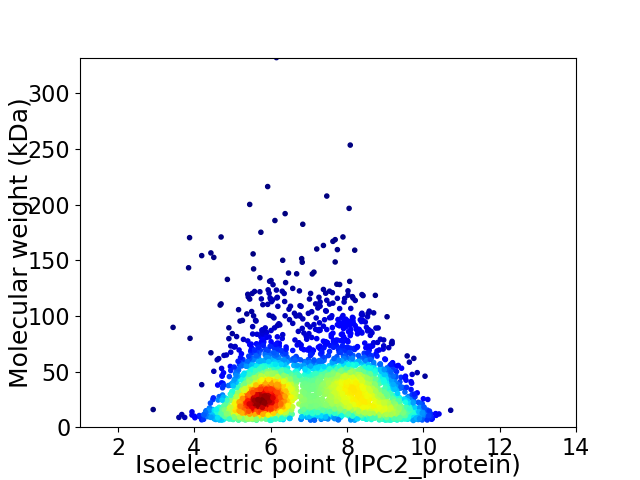
Virtual 2D-PAGE plot for 3394 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A177QTI2|A0A177QTI2_9PLAN DUF2294 domain-containing protein OS=Planctomycetaceae bacterium SCGC AG-212-F19 OX=1799658 GN=AYO44_05870 PE=4 SV=1
MM1 pKa = 7.47SANFSWWRR9 pKa = 11.84AGRR12 pKa = 11.84RR13 pKa = 11.84LWNSSVGDD21 pKa = 3.55WLQTGFGAARR31 pKa = 11.84SAPVRR36 pKa = 11.84RR37 pKa = 11.84GKK39 pKa = 10.41GRR41 pKa = 11.84RR42 pKa = 11.84SFRR45 pKa = 11.84PCLEE49 pKa = 3.89MLEE52 pKa = 4.94DD53 pKa = 3.72RR54 pKa = 11.84ALPSASWRR62 pKa = 11.84DD63 pKa = 3.67YY64 pKa = 11.68AHH66 pKa = 7.58DD67 pKa = 4.26PQHH70 pKa = 6.33TGVADD75 pKa = 3.7VASQPLEE82 pKa = 3.94QIHH85 pKa = 5.88WQTPVDD91 pKa = 4.67LNPQFSGSDD100 pKa = 3.51LLIHH104 pKa = 6.29YY105 pKa = 7.67GSPLVTANNTVIVPVKK121 pKa = 9.75TGAADD126 pKa = 3.46GFEE129 pKa = 4.09IEE131 pKa = 5.38GINAATGALKK141 pKa = 8.77WTVTSDD147 pKa = 4.35YY148 pKa = 10.82ILPPHH153 pKa = 6.56NWTPSYY159 pKa = 11.15SPALAPDD166 pKa = 3.19GRR168 pKa = 11.84LYY170 pKa = 10.73FAGVGGTIYY179 pKa = 11.09YY180 pKa = 9.83IDD182 pKa = 4.21NPDD185 pKa = 3.43ANGATVSGHH194 pKa = 4.79IAFYY198 pKa = 10.87GIGNYY203 pKa = 9.69NSSLDD208 pKa = 3.31GTVFINTPLTVDD220 pKa = 3.3SAGNLYY226 pKa = 10.65FGFEE230 pKa = 4.25VTGGNSLGLQSGIARR245 pKa = 11.84IGADD249 pKa = 3.8GTGTWIAASTAAGDD263 pKa = 3.86GNIVKK268 pKa = 9.67VVHH271 pKa = 6.24NCAPALSNDD280 pKa = 3.08GSSLYY285 pKa = 10.66VAVSTGSFGAGYY297 pKa = 10.02LVEE300 pKa = 5.2LNSTTLAPMARR311 pKa = 11.84VLLRR315 pKa = 11.84EE316 pKa = 4.03PLAGNAALLPDD327 pKa = 5.42DD328 pKa = 5.49GSASPTVGPDD338 pKa = 2.68GDD340 pKa = 4.06VYY342 pKa = 11.08FGVLEE347 pKa = 4.13NPFNSSKK354 pKa = 10.64GWLLHH359 pKa = 6.03FSGDD363 pKa = 3.36LSQTKK368 pKa = 8.96TPGAFGWDD376 pKa = 3.4DD377 pKa = 3.45TASIVAASLVPSYY390 pKa = 10.22HH391 pKa = 6.51GSSAYY396 pKa = 11.31LLMTKK401 pKa = 10.09YY402 pKa = 10.98NNYY405 pKa = 10.45AGLGGDD411 pKa = 5.21GINKK415 pKa = 9.41IAILDD420 pKa = 3.92PNATQTDD427 pKa = 4.13PRR429 pKa = 11.84TGATVMQEE437 pKa = 4.29VITIAGQTPDD447 pKa = 4.19PDD449 pKa = 4.02FPGNPGAVRR458 pKa = 11.84EE459 pKa = 4.28WCINTAAVDD468 pKa = 3.96PFTDD472 pKa = 3.4SVVANSEE479 pKa = 4.02DD480 pKa = 3.29GKK482 pKa = 10.93LYY484 pKa = 10.59RR485 pKa = 11.84WDD487 pKa = 3.61LATNTFSQVITLTTATGEE505 pKa = 4.32AYY507 pKa = 9.15TPTLIGPDD515 pKa = 3.37GTVYY519 pKa = 10.87AINRR523 pKa = 11.84ATLFAVGRR531 pKa = 11.84APSGPVTITGTGLFHH546 pKa = 7.56APAIYY551 pKa = 9.06PAGNTPDD558 pKa = 5.14SIAQGDD564 pKa = 3.98FNGDD568 pKa = 3.51GKK570 pKa = 10.22PDD572 pKa = 3.45LVVTNFFGGNVSVLINQGNGTFAAPVHH599 pKa = 5.96YY600 pKa = 8.75ATGSGPNGFVVADD613 pKa = 4.74FNHH616 pKa = 7.07DD617 pKa = 3.34GKK619 pKa = 11.28ADD621 pKa = 3.45IAVGNYY627 pKa = 8.84YY628 pKa = 11.01DD629 pKa = 3.78NAVSVLLGNGDD640 pKa = 3.71GTFQAAVNYY649 pKa = 10.29AVGSGPANEE658 pKa = 4.25MAFGDD663 pKa = 4.15FNGDD667 pKa = 3.24GNLDD671 pKa = 3.65LATANQNDD679 pKa = 4.1GTVSVLLGNGNGTFQPAVTYY699 pKa = 10.4AAGPGSDD706 pKa = 4.29GIVAADD712 pKa = 3.81LNGDD716 pKa = 3.83GKK718 pKa = 11.32LDD720 pKa = 3.8LAVANYY726 pKa = 9.85SGNNISVLLNQGNGTFAAPVTYY748 pKa = 10.57NVGANPWYY756 pKa = 9.95IEE758 pKa = 4.21KK759 pKa = 10.94GDD761 pKa = 3.92FNHH764 pKa = 7.56DD765 pKa = 3.38GTIDD769 pKa = 5.69LITADD774 pKa = 3.9SGGGTVSVLRR784 pKa = 11.84GNGDD788 pKa = 3.25GSFAAAVQYY797 pKa = 9.75TVGGFPDD804 pKa = 5.7GIAVADD810 pKa = 4.2LNGDD814 pKa = 3.77GNLDD818 pKa = 3.82LAVSNYY824 pKa = 7.25STNDD828 pKa = 2.98ASVLFGNADD837 pKa = 3.44GTFQAARR844 pKa = 11.84SVDD847 pKa = 3.34AGAGSGPNGITAADD861 pKa = 3.8FNGDD865 pKa = 3.4GKK867 pKa = 10.61IDD869 pKa = 3.66IASADD874 pKa = 3.75QNAGASVMLSGIVSAVEE891 pKa = 4.12GQSFTGVVASFTTSEE906 pKa = 4.42SNPSASEE913 pKa = 3.83YY914 pKa = 10.19SASINWGDD922 pKa = 3.75GNTSAGTIAADD933 pKa = 3.69GQGGFNVAGTHH944 pKa = 6.16TYY946 pKa = 10.92AEE948 pKa = 4.38EE949 pKa = 4.48GSYY952 pKa = 10.92SLSIVINGPGGASATVANVAGVADD976 pKa = 3.93PAVVVSGTSFTATEE990 pKa = 4.11GQAFSNQLVATFTDD1004 pKa = 3.54PGGAEE1009 pKa = 4.03SFAVSLGDD1017 pKa = 4.56PGFEE1021 pKa = 4.21TPNVGTGFFGAFQYY1035 pKa = 11.17DD1036 pKa = 3.64PTGAPWAYY1044 pKa = 10.49AGGAGVAGNQSGFTSGNPNAPQGTQVGFLQGPGGTISQTFTTGGGNYY1091 pKa = 9.71AVSFQAAQRR1100 pKa = 11.84GNYY1103 pKa = 8.42QPAGNQTIDD1112 pKa = 3.27VLIDD1116 pKa = 3.56GNVVGTFTPWGTNYY1130 pKa = 10.15QSFTTAPIALSGGTHH1145 pKa = 6.03TLTFEE1150 pKa = 4.46GLNSGDD1156 pKa = 3.11TTAFIDD1162 pKa = 3.84TVSIGGSGGGGPYY1175 pKa = 8.9TAAINWGDD1183 pKa = 3.64GSTSAADD1190 pKa = 3.64SIQYY1194 pKa = 10.22NATTGIFSVYY1204 pKa = 10.19GGHH1207 pKa = 6.72TYY1209 pKa = 11.21ASAAGSPLPVSVTIRR1224 pKa = 11.84HH1225 pKa = 5.72EE1226 pKa = 4.04NAPAATATSTATIQDD1241 pKa = 3.73VPLTPIDD1248 pKa = 4.1GPFTSPALYY1257 pKa = 10.33GAGSMPDD1264 pKa = 3.49SIATIRR1270 pKa = 11.84LDD1272 pKa = 3.46NGQVALLVANFGSANVTVLLPNGDD1296 pKa = 3.92GTFHH1300 pKa = 7.07SGGNLAVSNGPNGFAVGDD1318 pKa = 4.14FNHH1321 pKa = 7.36DD1322 pKa = 3.07GHH1324 pKa = 9.1LDD1326 pKa = 3.71FAVGNYY1332 pKa = 9.5YY1333 pKa = 10.89DD1334 pKa = 3.81STVTVYY1340 pKa = 10.81LGNGDD1345 pKa = 3.57GSFRR1349 pKa = 11.84SPAAYY1354 pKa = 9.75AVGANPANTMAVGDD1368 pKa = 4.05FNGDD1372 pKa = 3.29GNLDD1376 pKa = 3.69LATANQGNGTVTVLLGNSDD1395 pKa = 3.84GTFGSAVSYY1404 pKa = 11.07AGGSGNDD1411 pKa = 3.2SVVAVDD1417 pKa = 3.6VNGDD1421 pKa = 3.62GKK1423 pKa = 11.29LDD1425 pKa = 3.86LATANYY1431 pKa = 10.39NDD1433 pKa = 3.67GTASVLINQGNGTFAAPAIISVGSNPWYY1461 pKa = 10.03IDD1463 pKa = 3.2AGDD1466 pKa = 3.75FNKK1469 pKa = 10.42DD1470 pKa = 2.62GRR1472 pKa = 11.84MDD1474 pKa = 4.04LVVANLTGNSVSVLLNQGNGTFSRR1498 pKa = 11.84TDD1500 pKa = 3.31YY1501 pKa = 11.47GVGSWPTAVVVADD1514 pKa = 4.06MNRR1517 pKa = 11.84DD1518 pKa = 3.47GNPDD1522 pKa = 3.74LVVGNYY1528 pKa = 10.13GSTTVSVLLGNGDD1541 pKa = 3.76GTFQAALNFAAGGSGPNGVAVGDD1564 pKa = 3.92FNGDD1568 pKa = 3.44GKK1570 pKa = 10.21PDD1572 pKa = 3.48VAVADD1577 pKa = 3.94QDD1579 pKa = 3.94SGSASVLLNANLVSAVRR1596 pKa = 11.84NEE1598 pKa = 4.24SFTGAVGSFTDD1609 pKa = 4.6ANPLAPPTDD1618 pKa = 4.31FTATIHH1624 pKa = 6.22WGDD1627 pKa = 3.6GTTSTISASAFVRR1640 pKa = 11.84NAAGAFDD1647 pKa = 4.47VIGTHH1652 pKa = 7.02TYY1654 pKa = 8.35TQMGLFDD1661 pKa = 3.86ITIVIQDD1668 pKa = 3.6VGGATTTVSNLVSVHH1683 pKa = 5.63
MM1 pKa = 7.47SANFSWWRR9 pKa = 11.84AGRR12 pKa = 11.84RR13 pKa = 11.84LWNSSVGDD21 pKa = 3.55WLQTGFGAARR31 pKa = 11.84SAPVRR36 pKa = 11.84RR37 pKa = 11.84GKK39 pKa = 10.41GRR41 pKa = 11.84RR42 pKa = 11.84SFRR45 pKa = 11.84PCLEE49 pKa = 3.89MLEE52 pKa = 4.94DD53 pKa = 3.72RR54 pKa = 11.84ALPSASWRR62 pKa = 11.84DD63 pKa = 3.67YY64 pKa = 11.68AHH66 pKa = 7.58DD67 pKa = 4.26PQHH70 pKa = 6.33TGVADD75 pKa = 3.7VASQPLEE82 pKa = 3.94QIHH85 pKa = 5.88WQTPVDD91 pKa = 4.67LNPQFSGSDD100 pKa = 3.51LLIHH104 pKa = 6.29YY105 pKa = 7.67GSPLVTANNTVIVPVKK121 pKa = 9.75TGAADD126 pKa = 3.46GFEE129 pKa = 4.09IEE131 pKa = 5.38GINAATGALKK141 pKa = 8.77WTVTSDD147 pKa = 4.35YY148 pKa = 10.82ILPPHH153 pKa = 6.56NWTPSYY159 pKa = 11.15SPALAPDD166 pKa = 3.19GRR168 pKa = 11.84LYY170 pKa = 10.73FAGVGGTIYY179 pKa = 11.09YY180 pKa = 9.83IDD182 pKa = 4.21NPDD185 pKa = 3.43ANGATVSGHH194 pKa = 4.79IAFYY198 pKa = 10.87GIGNYY203 pKa = 9.69NSSLDD208 pKa = 3.31GTVFINTPLTVDD220 pKa = 3.3SAGNLYY226 pKa = 10.65FGFEE230 pKa = 4.25VTGGNSLGLQSGIARR245 pKa = 11.84IGADD249 pKa = 3.8GTGTWIAASTAAGDD263 pKa = 3.86GNIVKK268 pKa = 9.67VVHH271 pKa = 6.24NCAPALSNDD280 pKa = 3.08GSSLYY285 pKa = 10.66VAVSTGSFGAGYY297 pKa = 10.02LVEE300 pKa = 5.2LNSTTLAPMARR311 pKa = 11.84VLLRR315 pKa = 11.84EE316 pKa = 4.03PLAGNAALLPDD327 pKa = 5.42DD328 pKa = 5.49GSASPTVGPDD338 pKa = 2.68GDD340 pKa = 4.06VYY342 pKa = 11.08FGVLEE347 pKa = 4.13NPFNSSKK354 pKa = 10.64GWLLHH359 pKa = 6.03FSGDD363 pKa = 3.36LSQTKK368 pKa = 8.96TPGAFGWDD376 pKa = 3.4DD377 pKa = 3.45TASIVAASLVPSYY390 pKa = 10.22HH391 pKa = 6.51GSSAYY396 pKa = 11.31LLMTKK401 pKa = 10.09YY402 pKa = 10.98NNYY405 pKa = 10.45AGLGGDD411 pKa = 5.21GINKK415 pKa = 9.41IAILDD420 pKa = 3.92PNATQTDD427 pKa = 4.13PRR429 pKa = 11.84TGATVMQEE437 pKa = 4.29VITIAGQTPDD447 pKa = 4.19PDD449 pKa = 4.02FPGNPGAVRR458 pKa = 11.84EE459 pKa = 4.28WCINTAAVDD468 pKa = 3.96PFTDD472 pKa = 3.4SVVANSEE479 pKa = 4.02DD480 pKa = 3.29GKK482 pKa = 10.93LYY484 pKa = 10.59RR485 pKa = 11.84WDD487 pKa = 3.61LATNTFSQVITLTTATGEE505 pKa = 4.32AYY507 pKa = 9.15TPTLIGPDD515 pKa = 3.37GTVYY519 pKa = 10.87AINRR523 pKa = 11.84ATLFAVGRR531 pKa = 11.84APSGPVTITGTGLFHH546 pKa = 7.56APAIYY551 pKa = 9.06PAGNTPDD558 pKa = 5.14SIAQGDD564 pKa = 3.98FNGDD568 pKa = 3.51GKK570 pKa = 10.22PDD572 pKa = 3.45LVVTNFFGGNVSVLINQGNGTFAAPVHH599 pKa = 5.96YY600 pKa = 8.75ATGSGPNGFVVADD613 pKa = 4.74FNHH616 pKa = 7.07DD617 pKa = 3.34GKK619 pKa = 11.28ADD621 pKa = 3.45IAVGNYY627 pKa = 8.84YY628 pKa = 11.01DD629 pKa = 3.78NAVSVLLGNGDD640 pKa = 3.71GTFQAAVNYY649 pKa = 10.29AVGSGPANEE658 pKa = 4.25MAFGDD663 pKa = 4.15FNGDD667 pKa = 3.24GNLDD671 pKa = 3.65LATANQNDD679 pKa = 4.1GTVSVLLGNGNGTFQPAVTYY699 pKa = 10.4AAGPGSDD706 pKa = 4.29GIVAADD712 pKa = 3.81LNGDD716 pKa = 3.83GKK718 pKa = 11.32LDD720 pKa = 3.8LAVANYY726 pKa = 9.85SGNNISVLLNQGNGTFAAPVTYY748 pKa = 10.57NVGANPWYY756 pKa = 9.95IEE758 pKa = 4.21KK759 pKa = 10.94GDD761 pKa = 3.92FNHH764 pKa = 7.56DD765 pKa = 3.38GTIDD769 pKa = 5.69LITADD774 pKa = 3.9SGGGTVSVLRR784 pKa = 11.84GNGDD788 pKa = 3.25GSFAAAVQYY797 pKa = 9.75TVGGFPDD804 pKa = 5.7GIAVADD810 pKa = 4.2LNGDD814 pKa = 3.77GNLDD818 pKa = 3.82LAVSNYY824 pKa = 7.25STNDD828 pKa = 2.98ASVLFGNADD837 pKa = 3.44GTFQAARR844 pKa = 11.84SVDD847 pKa = 3.34AGAGSGPNGITAADD861 pKa = 3.8FNGDD865 pKa = 3.4GKK867 pKa = 10.61IDD869 pKa = 3.66IASADD874 pKa = 3.75QNAGASVMLSGIVSAVEE891 pKa = 4.12GQSFTGVVASFTTSEE906 pKa = 4.42SNPSASEE913 pKa = 3.83YY914 pKa = 10.19SASINWGDD922 pKa = 3.75GNTSAGTIAADD933 pKa = 3.69GQGGFNVAGTHH944 pKa = 6.16TYY946 pKa = 10.92AEE948 pKa = 4.38EE949 pKa = 4.48GSYY952 pKa = 10.92SLSIVINGPGGASATVANVAGVADD976 pKa = 3.93PAVVVSGTSFTATEE990 pKa = 4.11GQAFSNQLVATFTDD1004 pKa = 3.54PGGAEE1009 pKa = 4.03SFAVSLGDD1017 pKa = 4.56PGFEE1021 pKa = 4.21TPNVGTGFFGAFQYY1035 pKa = 11.17DD1036 pKa = 3.64PTGAPWAYY1044 pKa = 10.49AGGAGVAGNQSGFTSGNPNAPQGTQVGFLQGPGGTISQTFTTGGGNYY1091 pKa = 9.71AVSFQAAQRR1100 pKa = 11.84GNYY1103 pKa = 8.42QPAGNQTIDD1112 pKa = 3.27VLIDD1116 pKa = 3.56GNVVGTFTPWGTNYY1130 pKa = 10.15QSFTTAPIALSGGTHH1145 pKa = 6.03TLTFEE1150 pKa = 4.46GLNSGDD1156 pKa = 3.11TTAFIDD1162 pKa = 3.84TVSIGGSGGGGPYY1175 pKa = 8.9TAAINWGDD1183 pKa = 3.64GSTSAADD1190 pKa = 3.64SIQYY1194 pKa = 10.22NATTGIFSVYY1204 pKa = 10.19GGHH1207 pKa = 6.72TYY1209 pKa = 11.21ASAAGSPLPVSVTIRR1224 pKa = 11.84HH1225 pKa = 5.72EE1226 pKa = 4.04NAPAATATSTATIQDD1241 pKa = 3.73VPLTPIDD1248 pKa = 4.1GPFTSPALYY1257 pKa = 10.33GAGSMPDD1264 pKa = 3.49SIATIRR1270 pKa = 11.84LDD1272 pKa = 3.46NGQVALLVANFGSANVTVLLPNGDD1296 pKa = 3.92GTFHH1300 pKa = 7.07SGGNLAVSNGPNGFAVGDD1318 pKa = 4.14FNHH1321 pKa = 7.36DD1322 pKa = 3.07GHH1324 pKa = 9.1LDD1326 pKa = 3.71FAVGNYY1332 pKa = 9.5YY1333 pKa = 10.89DD1334 pKa = 3.81STVTVYY1340 pKa = 10.81LGNGDD1345 pKa = 3.57GSFRR1349 pKa = 11.84SPAAYY1354 pKa = 9.75AVGANPANTMAVGDD1368 pKa = 4.05FNGDD1372 pKa = 3.29GNLDD1376 pKa = 3.69LATANQGNGTVTVLLGNSDD1395 pKa = 3.84GTFGSAVSYY1404 pKa = 11.07AGGSGNDD1411 pKa = 3.2SVVAVDD1417 pKa = 3.6VNGDD1421 pKa = 3.62GKK1423 pKa = 11.29LDD1425 pKa = 3.86LATANYY1431 pKa = 10.39NDD1433 pKa = 3.67GTASVLINQGNGTFAAPAIISVGSNPWYY1461 pKa = 10.03IDD1463 pKa = 3.2AGDD1466 pKa = 3.75FNKK1469 pKa = 10.42DD1470 pKa = 2.62GRR1472 pKa = 11.84MDD1474 pKa = 4.04LVVANLTGNSVSVLLNQGNGTFSRR1498 pKa = 11.84TDD1500 pKa = 3.31YY1501 pKa = 11.47GVGSWPTAVVVADD1514 pKa = 4.06MNRR1517 pKa = 11.84DD1518 pKa = 3.47GNPDD1522 pKa = 3.74LVVGNYY1528 pKa = 10.13GSTTVSVLLGNGDD1541 pKa = 3.76GTFQAALNFAAGGSGPNGVAVGDD1564 pKa = 3.92FNGDD1568 pKa = 3.44GKK1570 pKa = 10.21PDD1572 pKa = 3.48VAVADD1577 pKa = 3.94QDD1579 pKa = 3.94SGSASVLLNANLVSAVRR1596 pKa = 11.84NEE1598 pKa = 4.24SFTGAVGSFTDD1609 pKa = 4.6ANPLAPPTDD1618 pKa = 4.31FTATIHH1624 pKa = 6.22WGDD1627 pKa = 3.6GTTSTISASAFVRR1640 pKa = 11.84NAAGAFDD1647 pKa = 4.47VIGTHH1652 pKa = 7.02TYY1654 pKa = 8.35TQMGLFDD1661 pKa = 3.86ITIVIQDD1668 pKa = 3.6VGGATTTVSNLVSVHH1683 pKa = 5.63
Molecular weight: 170.28 kDa
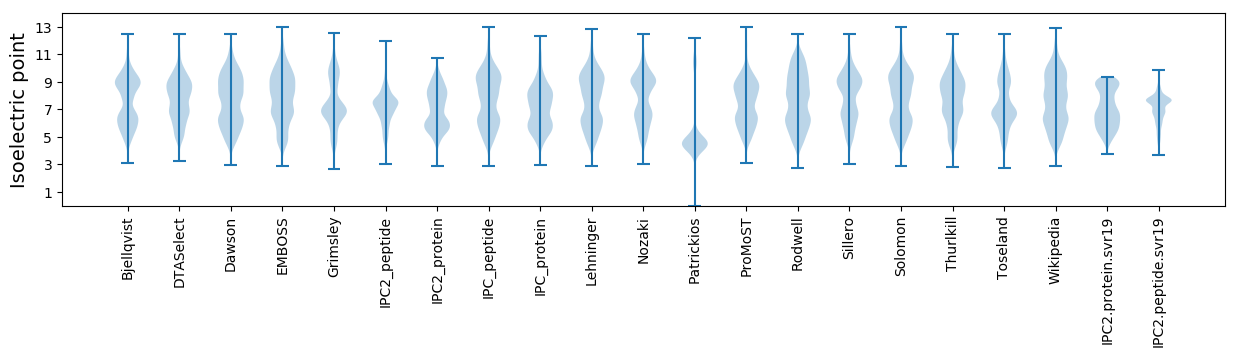
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A177QJR0|A0A177QJR0_9PLAN Methionine--tRNA ligase OS=Planctomycetaceae bacterium SCGC AG-212-F19 OX=1799658 GN=AYO44_08945 PE=3 SV=1
MM1 pKa = 7.82PAPPPVRR8 pKa = 11.84QAASDD13 pKa = 3.91RR14 pKa = 11.84SALACLRR21 pKa = 11.84MILASHH27 pKa = 6.33GRR29 pKa = 11.84SVTDD33 pKa = 4.27AVLQAPAPIAPGGIPVSDD51 pKa = 4.08LALLAGQFGLEE62 pKa = 4.0AEE64 pKa = 4.14IGTFNLAALTEE75 pKa = 4.1LLEE78 pKa = 4.74AGTYY82 pKa = 9.49PIVYY86 pKa = 9.9LNRR89 pKa = 11.84VHH91 pKa = 7.74LDD93 pKa = 3.04DD94 pKa = 3.48RR95 pKa = 11.84TIPRR99 pKa = 11.84KK100 pKa = 9.26LASRR104 pKa = 11.84RR105 pKa = 11.84CVVHH109 pKa = 6.52AVVPTRR115 pKa = 11.84LSSGFVWLNDD125 pKa = 3.4PRR127 pKa = 11.84SGTLRR132 pKa = 11.84KK133 pKa = 9.18VARR136 pKa = 11.84KK137 pKa = 9.58KK138 pKa = 10.73FEE140 pKa = 4.09AGRR143 pKa = 11.84RR144 pKa = 11.84DD145 pKa = 3.46LSNWCIVCRR154 pKa = 11.84RR155 pKa = 11.84AGRR158 pKa = 11.84GG159 pKa = 3.19
MM1 pKa = 7.82PAPPPVRR8 pKa = 11.84QAASDD13 pKa = 3.91RR14 pKa = 11.84SALACLRR21 pKa = 11.84MILASHH27 pKa = 6.33GRR29 pKa = 11.84SVTDD33 pKa = 4.27AVLQAPAPIAPGGIPVSDD51 pKa = 4.08LALLAGQFGLEE62 pKa = 4.0AEE64 pKa = 4.14IGTFNLAALTEE75 pKa = 4.1LLEE78 pKa = 4.74AGTYY82 pKa = 9.49PIVYY86 pKa = 9.9LNRR89 pKa = 11.84VHH91 pKa = 7.74LDD93 pKa = 3.04DD94 pKa = 3.48RR95 pKa = 11.84TIPRR99 pKa = 11.84KK100 pKa = 9.26LASRR104 pKa = 11.84RR105 pKa = 11.84CVVHH109 pKa = 6.52AVVPTRR115 pKa = 11.84LSSGFVWLNDD125 pKa = 3.4PRR127 pKa = 11.84SGTLRR132 pKa = 11.84KK133 pKa = 9.18VARR136 pKa = 11.84KK137 pKa = 9.58KK138 pKa = 10.73FEE140 pKa = 4.09AGRR143 pKa = 11.84RR144 pKa = 11.84DD145 pKa = 3.46LSNWCIVCRR154 pKa = 11.84RR155 pKa = 11.84AGRR158 pKa = 11.84GG159 pKa = 3.19
Molecular weight: 17.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1134279 |
58 |
2974 |
334.2 |
36.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.145 ± 0.056 | 1.142 ± 0.015 |
5.592 ± 0.031 | 5.566 ± 0.039 |
3.592 ± 0.024 | 8.218 ± 0.046 |
2.31 ± 0.019 | 4.195 ± 0.029 |
4.299 ± 0.046 | 10.461 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.008 ± 0.02 | 2.737 ± 0.032 |
6.16 ± 0.039 | 3.863 ± 0.028 |
7.068 ± 0.039 | 4.727 ± 0.032 |
5.386 ± 0.04 | 7.453 ± 0.034 |
1.673 ± 0.016 | 2.404 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
