
Geminocystis sp. NIES-3708
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Chroococcales; Chroococcaceae; Geminocystis; unclassified Geminocystis
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
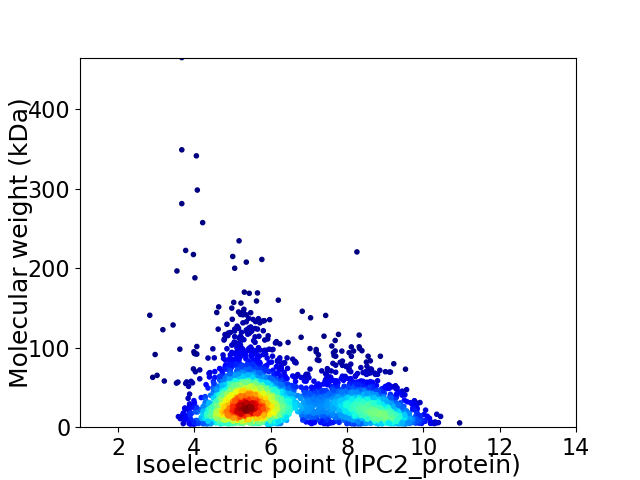
Virtual 2D-PAGE plot for 3607 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
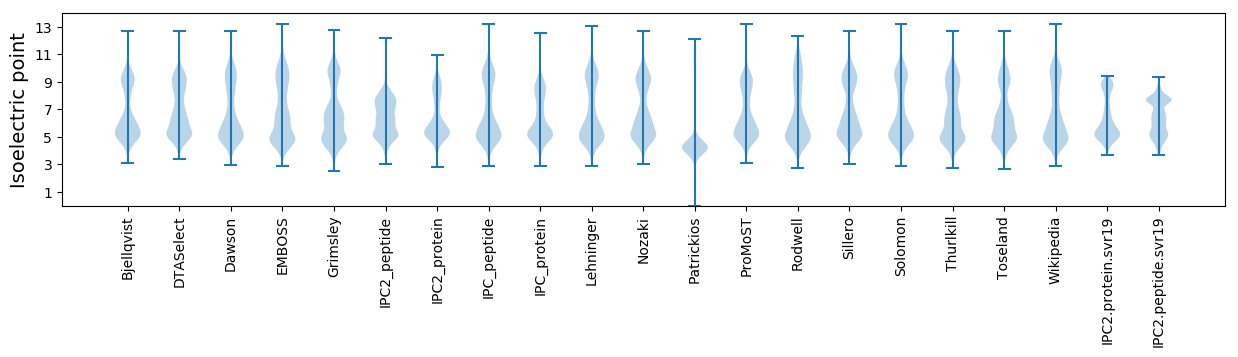
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D6AHQ5|A0A0D6AHQ5_9CHRO Uncharacterized protein OS=Geminocystis sp. NIES-3708 OX=1615909 GN=GM3708_2744 PE=4 SV=1
MM1 pKa = 7.83DD2 pKa = 4.04VKK4 pKa = 11.11NSVNIEE10 pKa = 4.05SPIATTKK17 pKa = 8.14VTPVSADD24 pKa = 3.43TSEE27 pKa = 4.04LTADD31 pKa = 4.53LSQKK35 pKa = 10.58LSLPEE40 pKa = 3.89VHH42 pKa = 7.05SSSTPLIKK50 pKa = 10.6NSLSSIISRR59 pKa = 11.84GISPPEE65 pKa = 3.79TVIKK69 pKa = 10.53LQDD72 pKa = 3.35SSPTFFSPIKK82 pKa = 9.28PLQAISAPSGDD93 pKa = 3.63YY94 pKa = 10.82RR95 pKa = 11.84IDD97 pKa = 3.56ALLGGNKK104 pKa = 8.99WGITNLTYY112 pKa = 10.78SFYY115 pKa = 11.15AGGSFYY121 pKa = 11.01GSEE124 pKa = 4.02TGLASVSEE132 pKa = 4.1AVKK135 pKa = 11.08NNVRR139 pKa = 11.84YY140 pKa = 9.78ILNNLIAPLINLTFTEE156 pKa = 4.7VTDD159 pKa = 3.71SPSNYY164 pKa = 9.29GQLRR168 pKa = 11.84YY169 pKa = 9.86LLSTDD174 pKa = 2.96PGYY177 pKa = 11.34AYY179 pKa = 10.27AYY181 pKa = 10.37YY182 pKa = 10.14PFSTDD187 pKa = 2.99TNQGNGNDD195 pKa = 3.51VAGDD199 pKa = 3.49VFLNPDD205 pKa = 3.31YY206 pKa = 11.66DD207 pKa = 4.47NNFDD211 pKa = 3.94TNGFRR216 pKa = 11.84GGAGTHH222 pKa = 7.06GYY224 pKa = 10.09QSLIHH229 pKa = 5.93EE230 pKa = 4.66TLHH233 pKa = 6.8ALGIKK238 pKa = 10.33HH239 pKa = 6.3SGNYY243 pKa = 8.97NGSGAGDD250 pKa = 4.05PPYY253 pKa = 10.79LPYY256 pKa = 11.04GEE258 pKa = 5.24EE259 pKa = 3.89NWDD262 pKa = 3.75NTLMTYY268 pKa = 10.75NFDD271 pKa = 4.14SGDD274 pKa = 3.75EE275 pKa = 4.17PSTPMAYY282 pKa = 10.1DD283 pKa = 3.71ILALQYY289 pKa = 11.03LYY291 pKa = 10.97GAKK294 pKa = 9.98SYY296 pKa = 11.33NSNNTIYY303 pKa = 10.29TFSDD307 pKa = 2.92TDD309 pKa = 4.01LYY311 pKa = 11.54SDD313 pKa = 3.94GSKK316 pKa = 9.33TVGSSTLNNKK326 pKa = 7.31ITIWDD331 pKa = 3.7SGGTDD336 pKa = 2.81ILNFTGLGSNTGGYY350 pKa = 9.63RR351 pKa = 11.84FDD353 pKa = 4.14MNPGGWFSTQTAFNGTGYY371 pKa = 10.62NVDD374 pKa = 3.86PGPPVSFSSGTTYY387 pKa = 10.92RR388 pKa = 11.84ATTNGTRR395 pKa = 11.84LGYY398 pKa = 10.56GIIIEE403 pKa = 4.34HH404 pKa = 6.67LVNSSSNDD412 pKa = 3.39YY413 pKa = 10.78IIANSAANQFSGYY426 pKa = 10.65GVITSTGNDD435 pKa = 3.54TIEE438 pKa = 4.82GGNSLDD444 pKa = 3.76TLDD447 pKa = 4.67LSEE450 pKa = 4.53FTSSSVSQIQSGNNLVLGLGSARR473 pKa = 11.84SITVKK478 pKa = 10.42DD479 pKa = 3.91YY480 pKa = 11.35YY481 pKa = 10.44LTSTDD486 pKa = 3.3RR487 pKa = 11.84LNIQFSSTNILTSFAIAATNANQTEE512 pKa = 4.72GNSGTKK518 pKa = 10.55AFTFTINRR526 pKa = 11.84SGTTTGANNVNWAVTGSGTNPANATDD552 pKa = 4.05FSGGVLPSGTVSFAANEE569 pKa = 4.05TSKK572 pKa = 11.28VITVNVQGDD581 pKa = 4.02TTVEE585 pKa = 3.62PDD587 pKa = 3.19EE588 pKa = 4.78NFTVTLSNATNGATITTATATGTILNDD615 pKa = 4.0DD616 pKa = 4.12GTNGNTFSNTSTISIPEE633 pKa = 3.9IGPASSYY640 pKa = 10.69PSTINVSGLSGNISKK655 pKa = 8.1VTVTLNNLSHH665 pKa = 7.06TFPDD669 pKa = 4.29DD670 pKa = 3.91LDD672 pKa = 4.16ILLVSPTGAKK682 pKa = 8.67TLLMSDD688 pKa = 3.3VGEE691 pKa = 4.58SNDD694 pKa = 4.1LSDD697 pKa = 3.55VTLTFDD703 pKa = 3.31PTATNFLPDD712 pKa = 3.46SGLITSGTYY721 pKa = 10.22KK722 pKa = 9.72PTDD725 pKa = 3.79FEE727 pKa = 5.59SDD729 pKa = 3.73DD730 pKa = 4.38VFDD733 pKa = 4.31SPAPVGPYY741 pKa = 10.04NADD744 pKa = 2.87LSVFNNTNPNGEE756 pKa = 3.64WRR758 pKa = 11.84LFVVDD763 pKa = 5.95DD764 pKa = 3.74ISGDD768 pKa = 3.49FGSIAGGLSLTFEE781 pKa = 4.54TTASFPTTLAIAATNANQTEE801 pKa = 4.72GNSGTKK807 pKa = 10.46AFTFTVTRR815 pKa = 11.84SGTTTGANNVNWAVTGSGTNPANATDD841 pKa = 4.05FSGGVLPSGTVSFAANEE858 pKa = 4.05TSKK861 pKa = 11.28VITVNVQGDD870 pKa = 4.02TTVEE874 pKa = 3.62PDD876 pKa = 3.19EE877 pKa = 4.78NFTVTLSNATNGATITTATAIGTIQNDD904 pKa = 3.71DD905 pKa = 3.67TPIPTNLAIAATNANQTEE923 pKa = 4.72GNSGTKK929 pKa = 10.46AFTFTVTRR937 pKa = 11.84SGTTTGANNVNWAVTGSGTNPANATDD963 pKa = 4.05FSGGVLPSGTVSFAANEE980 pKa = 4.05TSKK983 pKa = 11.28VITVNVQGDD992 pKa = 4.02TTVEE996 pKa = 3.62PDD998 pKa = 3.19EE999 pKa = 4.78NFTVTLSNATNGATITTATAIGTIQNDD1026 pKa = 3.7DD1027 pKa = 3.9TNTFLPQISISPNQTVIEE1045 pKa = 4.51GLTSLQNVTYY1055 pKa = 9.51TVILSQASSQTVTVQYY1071 pKa = 9.68ATTNGTAIAGSDD1083 pKa = 3.5YY1084 pKa = 10.42TATSGTLTFSPGQISKK1100 pKa = 9.28TIILPILNDD1109 pKa = 3.7FVNEE1113 pKa = 3.57FDD1115 pKa = 4.37EE1116 pKa = 4.8NFNLTLSNSTNANLSNSGVTTTIADD1141 pKa = 3.89TLFTAVTTTLSANVEE1156 pKa = 4.04NLILTGITAINGTGNGGNNIISGNTANNNLNGGAGNDD1193 pKa = 3.86TLNDD1197 pKa = 3.51GDD1199 pKa = 5.48GNDD1202 pKa = 4.26TLDD1205 pKa = 3.89GGAGSDD1211 pKa = 3.76VMSGGLGNDD1220 pKa = 4.16FYY1222 pKa = 11.6YY1223 pKa = 11.27VNSTVDD1229 pKa = 3.38VVVEE1233 pKa = 4.06NASQGTDD1240 pKa = 3.14TVISSITHH1248 pKa = 5.34TLAANVEE1255 pKa = 4.22NLTLIDD1261 pKa = 3.59ISAINGTGNTLNNTITGNIANNNLNGGDD1289 pKa = 4.06GNDD1292 pKa = 3.73TLTGGDD1298 pKa = 4.5GNDD1301 pKa = 3.66TLNGGTGSDD1310 pKa = 3.65RR1311 pKa = 11.84LTGGTGNDD1319 pKa = 3.86LYY1321 pKa = 11.51LVDD1324 pKa = 4.05NVGDD1328 pKa = 3.88IVSEE1332 pKa = 4.18TSTLATEE1339 pKa = 4.29IDD1341 pKa = 4.11TVQSSITYY1349 pKa = 8.19TLTTNVEE1356 pKa = 4.04NLTLTGTSAINGTGNTLNNTITGNIANNNLNGGDD1390 pKa = 4.06GNDD1393 pKa = 3.73TLTGGDD1399 pKa = 4.5GNDD1402 pKa = 3.82TLNGSIGSDD1411 pKa = 2.86RR1412 pKa = 11.84LTGGTGNDD1420 pKa = 3.8LYY1422 pKa = 10.73IVDD1425 pKa = 4.13NLGDD1429 pKa = 3.75IVIEE1433 pKa = 4.14TSTLATEE1440 pKa = 4.29IDD1442 pKa = 4.11TVQSLITYY1450 pKa = 7.52TLTTNVEE1457 pKa = 4.04NLTLTGTSAINGTGNILNNTITGNIANNNLNGGDD1491 pKa = 4.06GNDD1494 pKa = 3.73TLTGGDD1500 pKa = 4.5GNDD1503 pKa = 3.66TLNGGTGSDD1512 pKa = 3.57RR1513 pKa = 11.84LTSGTGNDD1521 pKa = 4.28LYY1523 pKa = 10.73IVDD1526 pKa = 4.08NVGDD1530 pKa = 3.96IVIEE1534 pKa = 4.12TSTLATEE1541 pKa = 4.29IDD1543 pKa = 4.11TVQSSITYY1551 pKa = 8.19TLTTNVEE1558 pKa = 4.04NLTLTGTSAINGTGNTLNNTITGNIANNNLNGGDD1592 pKa = 4.06GNDD1595 pKa = 3.73TLTGGDD1601 pKa = 4.5GNDD1604 pKa = 3.66TLNGGTGSDD1613 pKa = 3.65RR1614 pKa = 11.84LTGGTGNDD1622 pKa = 3.82LYY1624 pKa = 10.77IVDD1627 pKa = 4.07NVGDD1631 pKa = 4.01IVSEE1635 pKa = 4.18TSTLATEE1642 pKa = 4.29IDD1644 pKa = 4.11TVQSSITYY1652 pKa = 8.19TLTTNVEE1659 pKa = 4.04NLTLTGTTAINGTGNTLNNTITGNTANNSLDD1690 pKa = 3.41GGAGNDD1696 pKa = 3.81TLNGGTGIDD1705 pKa = 3.78SLIGGTGNDD1714 pKa = 4.13LYY1716 pKa = 10.77IVDD1719 pKa = 4.07NVGDD1723 pKa = 4.01IVSEE1727 pKa = 4.18TSTLATEE1734 pKa = 4.29IDD1736 pKa = 4.11TVQSSITYY1744 pKa = 8.16TLTDD1748 pKa = 3.28NVEE1751 pKa = 3.93NLTLTGTTAINGTGNTLKK1769 pKa = 10.87NVITGNSGNNSLSGGDD1785 pKa = 4.19GNDD1788 pKa = 3.4TLIGGDD1794 pKa = 4.26GNDD1797 pKa = 3.55TLIGGGSNDD1806 pKa = 3.71TLTGGNGLDD1815 pKa = 3.18SFRR1818 pKa = 11.84FNSASEE1824 pKa = 4.15GVDD1827 pKa = 4.3FITDD1831 pKa = 3.66FTVADD1836 pKa = 3.5DD1837 pKa = 4.75TIRR1840 pKa = 11.84VLGSAFGGGLVAGTLSISQFTIGSSATTSSHH1871 pKa = 6.01RR1872 pKa = 11.84FFYY1875 pKa = 10.82NSSNGGLFFDD1885 pKa = 4.46VDD1887 pKa = 4.08GNGATSAVQFATLNISLTMTNADD1910 pKa = 2.73IVVII1914 pKa = 4.28
MM1 pKa = 7.83DD2 pKa = 4.04VKK4 pKa = 11.11NSVNIEE10 pKa = 4.05SPIATTKK17 pKa = 8.14VTPVSADD24 pKa = 3.43TSEE27 pKa = 4.04LTADD31 pKa = 4.53LSQKK35 pKa = 10.58LSLPEE40 pKa = 3.89VHH42 pKa = 7.05SSSTPLIKK50 pKa = 10.6NSLSSIISRR59 pKa = 11.84GISPPEE65 pKa = 3.79TVIKK69 pKa = 10.53LQDD72 pKa = 3.35SSPTFFSPIKK82 pKa = 9.28PLQAISAPSGDD93 pKa = 3.63YY94 pKa = 10.82RR95 pKa = 11.84IDD97 pKa = 3.56ALLGGNKK104 pKa = 8.99WGITNLTYY112 pKa = 10.78SFYY115 pKa = 11.15AGGSFYY121 pKa = 11.01GSEE124 pKa = 4.02TGLASVSEE132 pKa = 4.1AVKK135 pKa = 11.08NNVRR139 pKa = 11.84YY140 pKa = 9.78ILNNLIAPLINLTFTEE156 pKa = 4.7VTDD159 pKa = 3.71SPSNYY164 pKa = 9.29GQLRR168 pKa = 11.84YY169 pKa = 9.86LLSTDD174 pKa = 2.96PGYY177 pKa = 11.34AYY179 pKa = 10.27AYY181 pKa = 10.37YY182 pKa = 10.14PFSTDD187 pKa = 2.99TNQGNGNDD195 pKa = 3.51VAGDD199 pKa = 3.49VFLNPDD205 pKa = 3.31YY206 pKa = 11.66DD207 pKa = 4.47NNFDD211 pKa = 3.94TNGFRR216 pKa = 11.84GGAGTHH222 pKa = 7.06GYY224 pKa = 10.09QSLIHH229 pKa = 5.93EE230 pKa = 4.66TLHH233 pKa = 6.8ALGIKK238 pKa = 10.33HH239 pKa = 6.3SGNYY243 pKa = 8.97NGSGAGDD250 pKa = 4.05PPYY253 pKa = 10.79LPYY256 pKa = 11.04GEE258 pKa = 5.24EE259 pKa = 3.89NWDD262 pKa = 3.75NTLMTYY268 pKa = 10.75NFDD271 pKa = 4.14SGDD274 pKa = 3.75EE275 pKa = 4.17PSTPMAYY282 pKa = 10.1DD283 pKa = 3.71ILALQYY289 pKa = 11.03LYY291 pKa = 10.97GAKK294 pKa = 9.98SYY296 pKa = 11.33NSNNTIYY303 pKa = 10.29TFSDD307 pKa = 2.92TDD309 pKa = 4.01LYY311 pKa = 11.54SDD313 pKa = 3.94GSKK316 pKa = 9.33TVGSSTLNNKK326 pKa = 7.31ITIWDD331 pKa = 3.7SGGTDD336 pKa = 2.81ILNFTGLGSNTGGYY350 pKa = 9.63RR351 pKa = 11.84FDD353 pKa = 4.14MNPGGWFSTQTAFNGTGYY371 pKa = 10.62NVDD374 pKa = 3.86PGPPVSFSSGTTYY387 pKa = 10.92RR388 pKa = 11.84ATTNGTRR395 pKa = 11.84LGYY398 pKa = 10.56GIIIEE403 pKa = 4.34HH404 pKa = 6.67LVNSSSNDD412 pKa = 3.39YY413 pKa = 10.78IIANSAANQFSGYY426 pKa = 10.65GVITSTGNDD435 pKa = 3.54TIEE438 pKa = 4.82GGNSLDD444 pKa = 3.76TLDD447 pKa = 4.67LSEE450 pKa = 4.53FTSSSVSQIQSGNNLVLGLGSARR473 pKa = 11.84SITVKK478 pKa = 10.42DD479 pKa = 3.91YY480 pKa = 11.35YY481 pKa = 10.44LTSTDD486 pKa = 3.3RR487 pKa = 11.84LNIQFSSTNILTSFAIAATNANQTEE512 pKa = 4.72GNSGTKK518 pKa = 10.55AFTFTINRR526 pKa = 11.84SGTTTGANNVNWAVTGSGTNPANATDD552 pKa = 4.05FSGGVLPSGTVSFAANEE569 pKa = 4.05TSKK572 pKa = 11.28VITVNVQGDD581 pKa = 4.02TTVEE585 pKa = 3.62PDD587 pKa = 3.19EE588 pKa = 4.78NFTVTLSNATNGATITTATATGTILNDD615 pKa = 4.0DD616 pKa = 4.12GTNGNTFSNTSTISIPEE633 pKa = 3.9IGPASSYY640 pKa = 10.69PSTINVSGLSGNISKK655 pKa = 8.1VTVTLNNLSHH665 pKa = 7.06TFPDD669 pKa = 4.29DD670 pKa = 3.91LDD672 pKa = 4.16ILLVSPTGAKK682 pKa = 8.67TLLMSDD688 pKa = 3.3VGEE691 pKa = 4.58SNDD694 pKa = 4.1LSDD697 pKa = 3.55VTLTFDD703 pKa = 3.31PTATNFLPDD712 pKa = 3.46SGLITSGTYY721 pKa = 10.22KK722 pKa = 9.72PTDD725 pKa = 3.79FEE727 pKa = 5.59SDD729 pKa = 3.73DD730 pKa = 4.38VFDD733 pKa = 4.31SPAPVGPYY741 pKa = 10.04NADD744 pKa = 2.87LSVFNNTNPNGEE756 pKa = 3.64WRR758 pKa = 11.84LFVVDD763 pKa = 5.95DD764 pKa = 3.74ISGDD768 pKa = 3.49FGSIAGGLSLTFEE781 pKa = 4.54TTASFPTTLAIAATNANQTEE801 pKa = 4.72GNSGTKK807 pKa = 10.46AFTFTVTRR815 pKa = 11.84SGTTTGANNVNWAVTGSGTNPANATDD841 pKa = 4.05FSGGVLPSGTVSFAANEE858 pKa = 4.05TSKK861 pKa = 11.28VITVNVQGDD870 pKa = 4.02TTVEE874 pKa = 3.62PDD876 pKa = 3.19EE877 pKa = 4.78NFTVTLSNATNGATITTATAIGTIQNDD904 pKa = 3.71DD905 pKa = 3.67TPIPTNLAIAATNANQTEE923 pKa = 4.72GNSGTKK929 pKa = 10.46AFTFTVTRR937 pKa = 11.84SGTTTGANNVNWAVTGSGTNPANATDD963 pKa = 4.05FSGGVLPSGTVSFAANEE980 pKa = 4.05TSKK983 pKa = 11.28VITVNVQGDD992 pKa = 4.02TTVEE996 pKa = 3.62PDD998 pKa = 3.19EE999 pKa = 4.78NFTVTLSNATNGATITTATAIGTIQNDD1026 pKa = 3.7DD1027 pKa = 3.9TNTFLPQISISPNQTVIEE1045 pKa = 4.51GLTSLQNVTYY1055 pKa = 9.51TVILSQASSQTVTVQYY1071 pKa = 9.68ATTNGTAIAGSDD1083 pKa = 3.5YY1084 pKa = 10.42TATSGTLTFSPGQISKK1100 pKa = 9.28TIILPILNDD1109 pKa = 3.7FVNEE1113 pKa = 3.57FDD1115 pKa = 4.37EE1116 pKa = 4.8NFNLTLSNSTNANLSNSGVTTTIADD1141 pKa = 3.89TLFTAVTTTLSANVEE1156 pKa = 4.04NLILTGITAINGTGNGGNNIISGNTANNNLNGGAGNDD1193 pKa = 3.86TLNDD1197 pKa = 3.51GDD1199 pKa = 5.48GNDD1202 pKa = 4.26TLDD1205 pKa = 3.89GGAGSDD1211 pKa = 3.76VMSGGLGNDD1220 pKa = 4.16FYY1222 pKa = 11.6YY1223 pKa = 11.27VNSTVDD1229 pKa = 3.38VVVEE1233 pKa = 4.06NASQGTDD1240 pKa = 3.14TVISSITHH1248 pKa = 5.34TLAANVEE1255 pKa = 4.22NLTLIDD1261 pKa = 3.59ISAINGTGNTLNNTITGNIANNNLNGGDD1289 pKa = 4.06GNDD1292 pKa = 3.73TLTGGDD1298 pKa = 4.5GNDD1301 pKa = 3.66TLNGGTGSDD1310 pKa = 3.65RR1311 pKa = 11.84LTGGTGNDD1319 pKa = 3.86LYY1321 pKa = 11.51LVDD1324 pKa = 4.05NVGDD1328 pKa = 3.88IVSEE1332 pKa = 4.18TSTLATEE1339 pKa = 4.29IDD1341 pKa = 4.11TVQSSITYY1349 pKa = 8.19TLTTNVEE1356 pKa = 4.04NLTLTGTSAINGTGNTLNNTITGNIANNNLNGGDD1390 pKa = 4.06GNDD1393 pKa = 3.73TLTGGDD1399 pKa = 4.5GNDD1402 pKa = 3.82TLNGSIGSDD1411 pKa = 2.86RR1412 pKa = 11.84LTGGTGNDD1420 pKa = 3.8LYY1422 pKa = 10.73IVDD1425 pKa = 4.13NLGDD1429 pKa = 3.75IVIEE1433 pKa = 4.14TSTLATEE1440 pKa = 4.29IDD1442 pKa = 4.11TVQSLITYY1450 pKa = 7.52TLTTNVEE1457 pKa = 4.04NLTLTGTSAINGTGNILNNTITGNIANNNLNGGDD1491 pKa = 4.06GNDD1494 pKa = 3.73TLTGGDD1500 pKa = 4.5GNDD1503 pKa = 3.66TLNGGTGSDD1512 pKa = 3.57RR1513 pKa = 11.84LTSGTGNDD1521 pKa = 4.28LYY1523 pKa = 10.73IVDD1526 pKa = 4.08NVGDD1530 pKa = 3.96IVIEE1534 pKa = 4.12TSTLATEE1541 pKa = 4.29IDD1543 pKa = 4.11TVQSSITYY1551 pKa = 8.19TLTTNVEE1558 pKa = 4.04NLTLTGTSAINGTGNTLNNTITGNIANNNLNGGDD1592 pKa = 4.06GNDD1595 pKa = 3.73TLTGGDD1601 pKa = 4.5GNDD1604 pKa = 3.66TLNGGTGSDD1613 pKa = 3.65RR1614 pKa = 11.84LTGGTGNDD1622 pKa = 3.82LYY1624 pKa = 10.77IVDD1627 pKa = 4.07NVGDD1631 pKa = 4.01IVSEE1635 pKa = 4.18TSTLATEE1642 pKa = 4.29IDD1644 pKa = 4.11TVQSSITYY1652 pKa = 8.19TLTTNVEE1659 pKa = 4.04NLTLTGTTAINGTGNTLNNTITGNTANNSLDD1690 pKa = 3.41GGAGNDD1696 pKa = 3.81TLNGGTGIDD1705 pKa = 3.78SLIGGTGNDD1714 pKa = 4.13LYY1716 pKa = 10.77IVDD1719 pKa = 4.07NVGDD1723 pKa = 4.01IVSEE1727 pKa = 4.18TSTLATEE1734 pKa = 4.29IDD1736 pKa = 4.11TVQSSITYY1744 pKa = 8.16TLTDD1748 pKa = 3.28NVEE1751 pKa = 3.93NLTLTGTTAINGTGNTLKK1769 pKa = 10.87NVITGNSGNNSLSGGDD1785 pKa = 4.19GNDD1788 pKa = 3.4TLIGGDD1794 pKa = 4.26GNDD1797 pKa = 3.55TLIGGGSNDD1806 pKa = 3.71TLTGGNGLDD1815 pKa = 3.18SFRR1818 pKa = 11.84FNSASEE1824 pKa = 4.15GVDD1827 pKa = 4.3FITDD1831 pKa = 3.66FTVADD1836 pKa = 3.5DD1837 pKa = 4.75TIRR1840 pKa = 11.84VLGSAFGGGLVAGTLSISQFTIGSSATTSSHH1871 pKa = 6.01RR1872 pKa = 11.84FFYY1875 pKa = 10.82NSSNGGLFFDD1885 pKa = 4.46VDD1887 pKa = 4.08GNGATSAVQFATLNISLTMTNADD1910 pKa = 2.73IVVII1914 pKa = 4.28
Molecular weight: 196.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D6AF49|A0A0D6AF49_9CHRO Uncharacterized protein OS=Geminocystis sp. NIES-3708 OX=1615909 GN=GM3708_1761 PE=4 SV=1
MM1 pKa = 7.64SKK3 pKa = 8.22HH4 pKa = 5.3TLNGTRR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 9.04QRR14 pKa = 11.84RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TEE26 pKa = 3.49NGRR29 pKa = 11.84KK30 pKa = 9.26VIQARR35 pKa = 11.84RR36 pKa = 11.84NKK38 pKa = 8.9GRR40 pKa = 11.84KK41 pKa = 8.69RR42 pKa = 11.84LSVV45 pKa = 3.2
MM1 pKa = 7.64SKK3 pKa = 8.22HH4 pKa = 5.3TLNGTRR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 9.04QRR14 pKa = 11.84RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TEE26 pKa = 3.49NGRR29 pKa = 11.84KK30 pKa = 9.26VIQARR35 pKa = 11.84RR36 pKa = 11.84NKK38 pKa = 8.9GRR40 pKa = 11.84KK41 pKa = 8.69RR42 pKa = 11.84LSVV45 pKa = 3.2
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1128004 |
37 |
4420 |
312.7 |
35.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.113 ± 0.051 | 0.95 ± 0.014 |
5.117 ± 0.038 | 6.574 ± 0.052 |
4.368 ± 0.029 | 6.166 ± 0.063 |
1.698 ± 0.023 | 8.826 ± 0.045 |
6.493 ± 0.052 | 10.771 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.802 ± 0.021 | 6.267 ± 0.067 |
3.915 ± 0.028 | 4.704 ± 0.029 |
3.867 ± 0.038 | 6.491 ± 0.036 |
5.549 ± 0.057 | 5.627 ± 0.042 |
1.269 ± 0.019 | 3.433 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
