
Amycolatopsis sulphurea
Taxonomy:
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
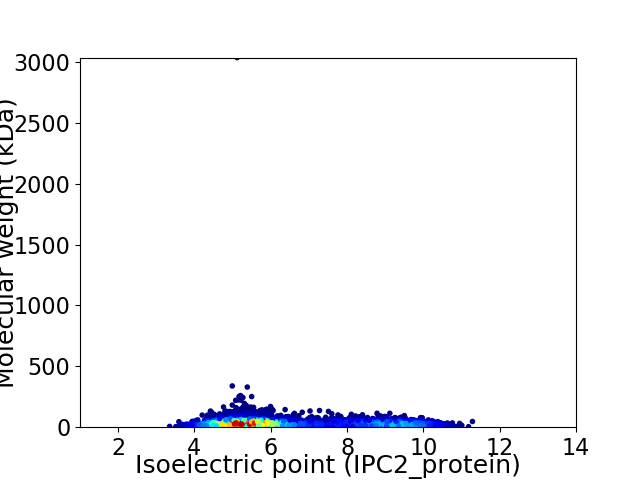
Virtual 2D-PAGE plot for 5941 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A9FAH0|A0A2A9FAH0_9PSEU Porphobilinogen deaminase OS=Amycolatopsis sulphurea OX=76022 GN=hemC PE=3 SV=1
MM1 pKa = 7.38WVDD4 pKa = 3.3EE5 pKa = 4.78SGGSDD10 pKa = 3.28ANGPEE15 pKa = 3.8AMTVHH20 pKa = 6.61VEE22 pKa = 4.03GQEE25 pKa = 3.96YY26 pKa = 8.97QAEE29 pKa = 4.32VNYY32 pKa = 10.8DD33 pKa = 3.47ADD35 pKa = 3.96HH36 pKa = 7.49DD37 pKa = 4.58GVADD41 pKa = 3.78TAVVEE46 pKa = 4.76HH47 pKa = 7.24GDD49 pKa = 3.32GTAQAFLDD57 pKa = 3.89TDD59 pKa = 3.62HH60 pKa = 7.47DD61 pKa = 4.63GVADD65 pKa = 4.51RR66 pKa = 11.84YY67 pKa = 9.85EE68 pKa = 3.86VLGASGQVQDD78 pKa = 3.53QAVYY82 pKa = 10.99DD83 pKa = 4.09EE84 pKa = 5.75ASGSWVEE91 pKa = 4.32SGGHH95 pKa = 5.75SGGTDD100 pKa = 2.92APAQNTSAGGHH111 pKa = 5.21IHH113 pKa = 7.4ADD115 pKa = 3.56LPGGGVDD122 pKa = 4.94AGAASIDD129 pKa = 3.84TDD131 pKa = 3.71HH132 pKa = 7.62DD133 pKa = 3.73GHH135 pKa = 6.79NDD137 pKa = 3.38TVVVEE142 pKa = 4.58TKK144 pKa = 10.63SGGTIAFTDD153 pKa = 3.55TDD155 pKa = 3.83GDD157 pKa = 3.94GAADD161 pKa = 3.5VAVEE165 pKa = 4.11IGSDD169 pKa = 3.6GSTSTYY175 pKa = 9.67EE176 pKa = 3.9HH177 pKa = 7.0TGPGQWSEE185 pKa = 4.05TSGPPDD191 pKa = 3.47RR192 pKa = 11.84AFDD195 pKa = 4.05PAADD199 pKa = 3.94AAWGGEE205 pKa = 4.04GTQLLNGVARR215 pKa = 11.84IDD217 pKa = 3.66SGTGQWISPNN227 pKa = 3.39
MM1 pKa = 7.38WVDD4 pKa = 3.3EE5 pKa = 4.78SGGSDD10 pKa = 3.28ANGPEE15 pKa = 3.8AMTVHH20 pKa = 6.61VEE22 pKa = 4.03GQEE25 pKa = 3.96YY26 pKa = 8.97QAEE29 pKa = 4.32VNYY32 pKa = 10.8DD33 pKa = 3.47ADD35 pKa = 3.96HH36 pKa = 7.49DD37 pKa = 4.58GVADD41 pKa = 3.78TAVVEE46 pKa = 4.76HH47 pKa = 7.24GDD49 pKa = 3.32GTAQAFLDD57 pKa = 3.89TDD59 pKa = 3.62HH60 pKa = 7.47DD61 pKa = 4.63GVADD65 pKa = 4.51RR66 pKa = 11.84YY67 pKa = 9.85EE68 pKa = 3.86VLGASGQVQDD78 pKa = 3.53QAVYY82 pKa = 10.99DD83 pKa = 4.09EE84 pKa = 5.75ASGSWVEE91 pKa = 4.32SGGHH95 pKa = 5.75SGGTDD100 pKa = 2.92APAQNTSAGGHH111 pKa = 5.21IHH113 pKa = 7.4ADD115 pKa = 3.56LPGGGVDD122 pKa = 4.94AGAASIDD129 pKa = 3.84TDD131 pKa = 3.71HH132 pKa = 7.62DD133 pKa = 3.73GHH135 pKa = 6.79NDD137 pKa = 3.38TVVVEE142 pKa = 4.58TKK144 pKa = 10.63SGGTIAFTDD153 pKa = 3.55TDD155 pKa = 3.83GDD157 pKa = 3.94GAADD161 pKa = 3.5VAVEE165 pKa = 4.11IGSDD169 pKa = 3.6GSTSTYY175 pKa = 9.67EE176 pKa = 3.9HH177 pKa = 7.0TGPGQWSEE185 pKa = 4.05TSGPPDD191 pKa = 3.47RR192 pKa = 11.84AFDD195 pKa = 4.05PAADD199 pKa = 3.94AAWGGEE205 pKa = 4.04GTQLLNGVARR215 pKa = 11.84IDD217 pKa = 3.66SGTGQWISPNN227 pKa = 3.39
Molecular weight: 22.93 kDa
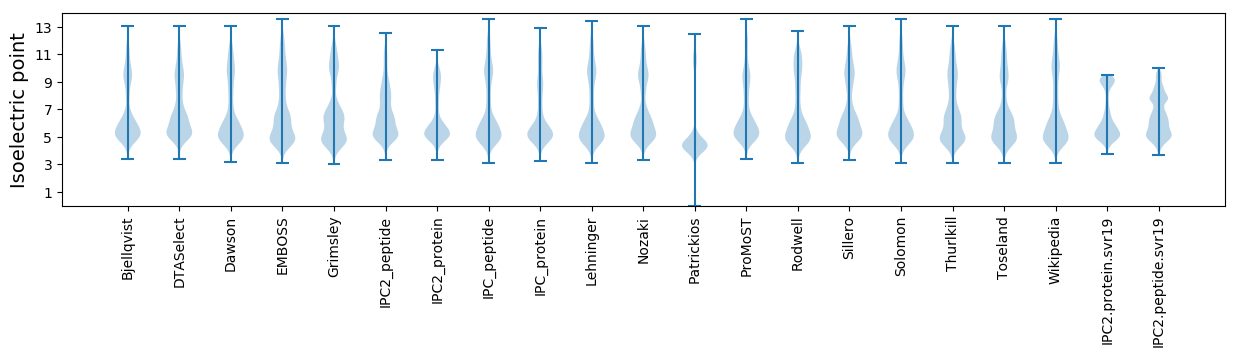
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A9FCR8|A0A2A9FCR8_9PSEU Crotonobetaine/carnitine-CoA ligase OS=Amycolatopsis sulphurea OX=76022 GN=ATK36_4367 PE=4 SV=1
MM1 pKa = 7.67PVPRR5 pKa = 11.84PWRR8 pKa = 11.84LHH10 pKa = 4.53SARR13 pKa = 11.84PGDD16 pKa = 3.56LHH18 pKa = 7.09RR19 pKa = 11.84WRR21 pKa = 11.84ARR23 pKa = 11.84PGPPAPAPPHH33 pKa = 5.85LVRR36 pKa = 11.84HH37 pKa = 5.78GRR39 pKa = 11.84RR40 pKa = 11.84TLRR43 pKa = 11.84PRR45 pKa = 11.84PVPRR49 pKa = 11.84GRR51 pKa = 11.84RR52 pKa = 11.84APAPCRR58 pKa = 11.84ARR60 pKa = 11.84RR61 pKa = 11.84GAAARR66 pKa = 11.84LHH68 pKa = 6.03SRR70 pKa = 11.84ARR72 pKa = 11.84PGTRR76 pKa = 11.84TLPVLRR82 pKa = 11.84SRR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84ASRR89 pKa = 11.84GGPQRR94 pKa = 11.84RR95 pKa = 11.84PVRR98 pKa = 11.84TGAEE102 pKa = 4.02LPRR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84QVVTRR112 pKa = 11.84RR113 pKa = 11.84ARR115 pKa = 11.84WAVILQAAARR125 pKa = 11.84GTRR128 pKa = 11.84LLMNPRR134 pKa = 11.84VAQLVALRR142 pKa = 11.84AWPVRR147 pKa = 11.84PLVRR151 pKa = 11.84WAPRR155 pKa = 11.84PAPPGRR161 pKa = 11.84GVVARR166 pKa = 11.84APPRR170 pKa = 11.84AGRR173 pKa = 11.84SRR175 pKa = 11.84IPATPRR181 pKa = 11.84VVPARR186 pKa = 11.84PRR188 pKa = 11.84LVIRR192 pKa = 11.84RR193 pKa = 11.84SRR195 pKa = 11.84RR196 pKa = 11.84PSIPQVFRR204 pKa = 11.84PLVRR208 pKa = 11.84AARR211 pKa = 11.84RR212 pKa = 11.84APTPSIRR219 pKa = 11.84QAVQLRR225 pKa = 11.84RR226 pKa = 11.84QVARR230 pKa = 11.84RR231 pKa = 11.84ARR233 pKa = 11.84TPPVPQAAQPPARR246 pKa = 11.84RR247 pKa = 11.84ATRR250 pKa = 11.84LQAGQRR256 pKa = 11.84PAVRR260 pKa = 11.84RR261 pKa = 11.84APRR264 pKa = 11.84SVQRR268 pKa = 11.84ARR270 pKa = 11.84LQVPRR275 pKa = 11.84PAVWVAPRR283 pKa = 11.84TAASPSRR290 pKa = 11.84PSPNPSAIRR299 pKa = 11.84ATSRR303 pKa = 11.84ATVRR307 pKa = 11.84SPVARR312 pKa = 11.84RR313 pKa = 11.84ARR315 pKa = 11.84SRR317 pKa = 11.84GPAAEE322 pKa = 4.04VRR324 pKa = 11.84RR325 pKa = 11.84SPRR328 pKa = 11.84RR329 pKa = 11.84VVSVGRR335 pKa = 11.84RR336 pKa = 11.84AGRR339 pKa = 11.84WRR341 pKa = 11.84PVVRR345 pKa = 11.84AGIRR349 pKa = 11.84AGGPGRR355 pKa = 11.84ARR357 pKa = 11.84RR358 pKa = 11.84RR359 pKa = 11.84CSGMRR364 pKa = 11.84RR365 pKa = 11.84GRR367 pKa = 11.84PAGPVPIPRR376 pKa = 11.84CRR378 pKa = 11.84SRR380 pKa = 11.84VSRR383 pKa = 11.84GRR385 pKa = 11.84PRR387 pKa = 11.84RR388 pKa = 11.84TSSVSTDD395 pKa = 2.9SQARR399 pKa = 11.84KK400 pKa = 8.1VWPVRR405 pKa = 11.84KK406 pKa = 9.7VRR408 pKa = 11.84AFRR411 pKa = 11.84QACQARR417 pKa = 11.84AAA419 pKa = 4.03
MM1 pKa = 7.67PVPRR5 pKa = 11.84PWRR8 pKa = 11.84LHH10 pKa = 4.53SARR13 pKa = 11.84PGDD16 pKa = 3.56LHH18 pKa = 7.09RR19 pKa = 11.84WRR21 pKa = 11.84ARR23 pKa = 11.84PGPPAPAPPHH33 pKa = 5.85LVRR36 pKa = 11.84HH37 pKa = 5.78GRR39 pKa = 11.84RR40 pKa = 11.84TLRR43 pKa = 11.84PRR45 pKa = 11.84PVPRR49 pKa = 11.84GRR51 pKa = 11.84RR52 pKa = 11.84APAPCRR58 pKa = 11.84ARR60 pKa = 11.84RR61 pKa = 11.84GAAARR66 pKa = 11.84LHH68 pKa = 6.03SRR70 pKa = 11.84ARR72 pKa = 11.84PGTRR76 pKa = 11.84TLPVLRR82 pKa = 11.84SRR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84ASRR89 pKa = 11.84GGPQRR94 pKa = 11.84RR95 pKa = 11.84PVRR98 pKa = 11.84TGAEE102 pKa = 4.02LPRR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84QVVTRR112 pKa = 11.84RR113 pKa = 11.84ARR115 pKa = 11.84WAVILQAAARR125 pKa = 11.84GTRR128 pKa = 11.84LLMNPRR134 pKa = 11.84VAQLVALRR142 pKa = 11.84AWPVRR147 pKa = 11.84PLVRR151 pKa = 11.84WAPRR155 pKa = 11.84PAPPGRR161 pKa = 11.84GVVARR166 pKa = 11.84APPRR170 pKa = 11.84AGRR173 pKa = 11.84SRR175 pKa = 11.84IPATPRR181 pKa = 11.84VVPARR186 pKa = 11.84PRR188 pKa = 11.84LVIRR192 pKa = 11.84RR193 pKa = 11.84SRR195 pKa = 11.84RR196 pKa = 11.84PSIPQVFRR204 pKa = 11.84PLVRR208 pKa = 11.84AARR211 pKa = 11.84RR212 pKa = 11.84APTPSIRR219 pKa = 11.84QAVQLRR225 pKa = 11.84RR226 pKa = 11.84QVARR230 pKa = 11.84RR231 pKa = 11.84ARR233 pKa = 11.84TPPVPQAAQPPARR246 pKa = 11.84RR247 pKa = 11.84ATRR250 pKa = 11.84LQAGQRR256 pKa = 11.84PAVRR260 pKa = 11.84RR261 pKa = 11.84APRR264 pKa = 11.84SVQRR268 pKa = 11.84ARR270 pKa = 11.84LQVPRR275 pKa = 11.84PAVWVAPRR283 pKa = 11.84TAASPSRR290 pKa = 11.84PSPNPSAIRR299 pKa = 11.84ATSRR303 pKa = 11.84ATVRR307 pKa = 11.84SPVARR312 pKa = 11.84RR313 pKa = 11.84ARR315 pKa = 11.84SRR317 pKa = 11.84GPAAEE322 pKa = 4.04VRR324 pKa = 11.84RR325 pKa = 11.84SPRR328 pKa = 11.84RR329 pKa = 11.84VVSVGRR335 pKa = 11.84RR336 pKa = 11.84AGRR339 pKa = 11.84WRR341 pKa = 11.84PVVRR345 pKa = 11.84AGIRR349 pKa = 11.84AGGPGRR355 pKa = 11.84ARR357 pKa = 11.84RR358 pKa = 11.84RR359 pKa = 11.84CSGMRR364 pKa = 11.84RR365 pKa = 11.84GRR367 pKa = 11.84PAGPVPIPRR376 pKa = 11.84CRR378 pKa = 11.84SRR380 pKa = 11.84VSRR383 pKa = 11.84GRR385 pKa = 11.84PRR387 pKa = 11.84RR388 pKa = 11.84TSSVSTDD395 pKa = 2.9SQARR399 pKa = 11.84KK400 pKa = 8.1VWPVRR405 pKa = 11.84KK406 pKa = 9.7VRR408 pKa = 11.84AFRR411 pKa = 11.84QACQARR417 pKa = 11.84AAA419 pKa = 4.03
Molecular weight: 46.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1913325 |
29 |
28289 |
322.1 |
34.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.273 ± 0.047 | 0.777 ± 0.014 |
5.795 ± 0.027 | 5.672 ± 0.037 |
2.866 ± 0.018 | 9.434 ± 0.033 |
2.262 ± 0.015 | 3.394 ± 0.026 |
2.211 ± 0.023 | 10.372 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.64 ± 0.014 | 1.872 ± 0.018 |
6.157 ± 0.035 | 2.959 ± 0.017 |
7.934 ± 0.036 | 5.254 ± 0.038 |
6.024 ± 0.027 | 8.646 ± 0.032 |
1.465 ± 0.012 | 1.993 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
