
Pacific flying fox faeces associated circular DNA virus-5
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
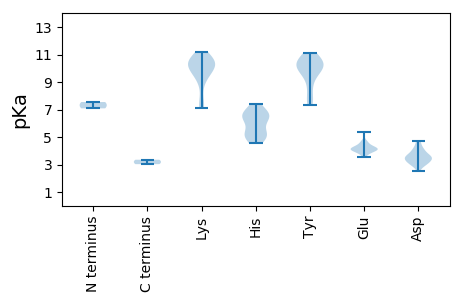
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A140CTT9|A0A140CTT9_9VIRU Putative capsid protein OS=Pacific flying fox faeces associated circular DNA virus-5 OX=1796014 PE=4 SV=1
MM1 pKa = 7.09GWRR4 pKa = 11.84RR5 pKa = 11.84YY6 pKa = 8.83HH7 pKa = 5.04RR8 pKa = 11.84HH9 pKa = 5.05SRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 7.64YY14 pKa = 8.06HH15 pKa = 7.42KK16 pKa = 10.45ARR18 pKa = 11.84QAYY21 pKa = 10.22DD22 pKa = 3.03NFAFFIPHH30 pKa = 5.19SVIYY34 pKa = 10.31LALQGNRR41 pKa = 11.84FPGGGEE47 pKa = 4.01PIPYY51 pKa = 9.4YY52 pKa = 10.04PRR54 pKa = 11.84VYY56 pKa = 11.1GNILNFTEE64 pKa = 4.72PYY66 pKa = 9.94IGISVTQGAPQLFYY80 pKa = 10.88FQLNDD85 pKa = 3.13IPNYY89 pKa = 10.39LQFTGLYY96 pKa = 8.14DD97 pKa = 2.99QYY99 pKa = 10.85RR100 pKa = 11.84ISRR103 pKa = 11.84ITASFFPSHH112 pKa = 6.48TSQPINNSAYY122 pKa = 9.74MKK124 pKa = 10.45TDD126 pKa = 3.48GTWHH130 pKa = 5.19QTSDD134 pKa = 3.57PDD136 pKa = 4.71AVTEE140 pKa = 4.05QYY142 pKa = 7.36TTGVSSHH149 pKa = 6.71SDD151 pKa = 2.5SGTYY155 pKa = 10.8YY156 pKa = 9.79NTFGDD161 pKa = 4.65FMMTCWVDD169 pKa = 3.21VDD171 pKa = 4.21GTNSSLTQSSYY182 pKa = 10.4QNVTQLHH189 pKa = 5.65KK190 pKa = 10.44WRR192 pKa = 11.84WTEE195 pKa = 3.57KK196 pKa = 10.73HH197 pKa = 6.28EE198 pKa = 4.17FTFWPKK204 pKa = 10.44PYY206 pKa = 10.14LVSANVGGTTGPAPDD221 pKa = 4.21LWQDD225 pKa = 3.53CSDD228 pKa = 4.06VDD230 pKa = 3.45IQYY233 pKa = 10.2WGLWVQALPQLPTGQLYY250 pKa = 10.2NQTQQYY256 pKa = 7.39DD257 pKa = 3.66TTNIMLYY264 pKa = 8.85CTYY267 pKa = 10.06HH268 pKa = 6.71VEE270 pKa = 4.33FRR272 pKa = 11.84FRR274 pKa = 11.84QQ275 pKa = 3.35
MM1 pKa = 7.09GWRR4 pKa = 11.84RR5 pKa = 11.84YY6 pKa = 8.83HH7 pKa = 5.04RR8 pKa = 11.84HH9 pKa = 5.05SRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 7.64YY14 pKa = 8.06HH15 pKa = 7.42KK16 pKa = 10.45ARR18 pKa = 11.84QAYY21 pKa = 10.22DD22 pKa = 3.03NFAFFIPHH30 pKa = 5.19SVIYY34 pKa = 10.31LALQGNRR41 pKa = 11.84FPGGGEE47 pKa = 4.01PIPYY51 pKa = 9.4YY52 pKa = 10.04PRR54 pKa = 11.84VYY56 pKa = 11.1GNILNFTEE64 pKa = 4.72PYY66 pKa = 9.94IGISVTQGAPQLFYY80 pKa = 10.88FQLNDD85 pKa = 3.13IPNYY89 pKa = 10.39LQFTGLYY96 pKa = 8.14DD97 pKa = 2.99QYY99 pKa = 10.85RR100 pKa = 11.84ISRR103 pKa = 11.84ITASFFPSHH112 pKa = 6.48TSQPINNSAYY122 pKa = 9.74MKK124 pKa = 10.45TDD126 pKa = 3.48GTWHH130 pKa = 5.19QTSDD134 pKa = 3.57PDD136 pKa = 4.71AVTEE140 pKa = 4.05QYY142 pKa = 7.36TTGVSSHH149 pKa = 6.71SDD151 pKa = 2.5SGTYY155 pKa = 10.8YY156 pKa = 9.79NTFGDD161 pKa = 4.65FMMTCWVDD169 pKa = 3.21VDD171 pKa = 4.21GTNSSLTQSSYY182 pKa = 10.4QNVTQLHH189 pKa = 5.65KK190 pKa = 10.44WRR192 pKa = 11.84WTEE195 pKa = 3.57KK196 pKa = 10.73HH197 pKa = 6.28EE198 pKa = 4.17FTFWPKK204 pKa = 10.44PYY206 pKa = 10.14LVSANVGGTTGPAPDD221 pKa = 4.21LWQDD225 pKa = 3.53CSDD228 pKa = 4.06VDD230 pKa = 3.45IQYY233 pKa = 10.2WGLWVQALPQLPTGQLYY250 pKa = 10.2NQTQQYY256 pKa = 7.39DD257 pKa = 3.66TTNIMLYY264 pKa = 8.85CTYY267 pKa = 10.06HH268 pKa = 6.71VEE270 pKa = 4.33FRR272 pKa = 11.84FRR274 pKa = 11.84QQ275 pKa = 3.35
Molecular weight: 32.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTT9|A0A140CTT9_9VIRU Putative capsid protein OS=Pacific flying fox faeces associated circular DNA virus-5 OX=1796014 PE=4 SV=1
MM1 pKa = 7.55GSRR4 pKa = 11.84RR5 pKa = 11.84YY6 pKa = 10.29VFTIFVSSDD15 pKa = 3.28RR16 pKa = 11.84DD17 pKa = 3.56WITEE21 pKa = 4.18GIEE24 pKa = 4.1RR25 pKa = 11.84QNVKK29 pKa = 10.72YY30 pKa = 10.41LVAQQEE36 pKa = 4.53ICSEE40 pKa = 4.15TGRR43 pKa = 11.84LHH45 pKa = 5.03WQGYY49 pKa = 9.76AEE51 pKa = 4.17FKK53 pKa = 10.1KK54 pKa = 10.48VCRR57 pKa = 11.84IPGAKK62 pKa = 9.8SILGCDD68 pKa = 3.51WAHH71 pKa = 6.33LAVPNGDD78 pKa = 2.94RR79 pKa = 11.84KK80 pKa = 10.79ANKK83 pKa = 9.88DD84 pKa = 3.61YY85 pKa = 9.52CTKK88 pKa = 10.29LASRR92 pKa = 11.84APGGEE97 pKa = 4.0SIEE100 pKa = 3.84IGKK103 pKa = 10.02FEE105 pKa = 4.68EE106 pKa = 5.35GGQGRR111 pKa = 11.84KK112 pKa = 7.1STSVAEE118 pKa = 4.06LRR120 pKa = 11.84EE121 pKa = 4.3EE122 pKa = 3.84IRR124 pKa = 11.84GGQTTLRR131 pKa = 11.84QEE133 pKa = 3.95IVEE136 pKa = 4.08GRR138 pKa = 11.84VLNIGAIRR146 pKa = 11.84YY147 pKa = 7.51FEE149 pKa = 4.32RR150 pKa = 11.84LQSEE154 pKa = 4.57LDD156 pKa = 3.06SGYY159 pKa = 10.62QRR161 pKa = 11.84GHH163 pKa = 6.19QIAAYY168 pKa = 10.4LFGTPGTGKK177 pKa = 9.75SHH179 pKa = 6.88IAHH182 pKa = 6.78AAVEE186 pKa = 4.09EE187 pKa = 4.26RR188 pKa = 11.84TWYY191 pKa = 10.97SKK193 pKa = 11.18DD194 pKa = 3.1ATTKK198 pKa = 8.77WWDD201 pKa = 3.79GYY203 pKa = 10.84SGEE206 pKa = 4.36EE207 pKa = 4.13VVIVDD212 pKa = 4.01DD213 pKa = 3.94WSGPSTQAEE222 pKa = 3.75ADD224 pKa = 3.42QWKK227 pKa = 9.48KK228 pKa = 9.3WVDD231 pKa = 3.54RR232 pKa = 11.84YY233 pKa = 9.54PCRR236 pKa = 11.84VEE238 pKa = 4.08TKK240 pKa = 10.41GGHH243 pKa = 4.53VWLKK247 pKa = 7.19TKK249 pKa = 9.64QWIFTSNVRR258 pKa = 11.84WVALPVFFDD267 pKa = 3.36AALKK271 pKa = 10.55RR272 pKa = 11.84RR273 pKa = 11.84FIGNGG278 pKa = 3.03
MM1 pKa = 7.55GSRR4 pKa = 11.84RR5 pKa = 11.84YY6 pKa = 10.29VFTIFVSSDD15 pKa = 3.28RR16 pKa = 11.84DD17 pKa = 3.56WITEE21 pKa = 4.18GIEE24 pKa = 4.1RR25 pKa = 11.84QNVKK29 pKa = 10.72YY30 pKa = 10.41LVAQQEE36 pKa = 4.53ICSEE40 pKa = 4.15TGRR43 pKa = 11.84LHH45 pKa = 5.03WQGYY49 pKa = 9.76AEE51 pKa = 4.17FKK53 pKa = 10.1KK54 pKa = 10.48VCRR57 pKa = 11.84IPGAKK62 pKa = 9.8SILGCDD68 pKa = 3.51WAHH71 pKa = 6.33LAVPNGDD78 pKa = 2.94RR79 pKa = 11.84KK80 pKa = 10.79ANKK83 pKa = 9.88DD84 pKa = 3.61YY85 pKa = 9.52CTKK88 pKa = 10.29LASRR92 pKa = 11.84APGGEE97 pKa = 4.0SIEE100 pKa = 3.84IGKK103 pKa = 10.02FEE105 pKa = 4.68EE106 pKa = 5.35GGQGRR111 pKa = 11.84KK112 pKa = 7.1STSVAEE118 pKa = 4.06LRR120 pKa = 11.84EE121 pKa = 4.3EE122 pKa = 3.84IRR124 pKa = 11.84GGQTTLRR131 pKa = 11.84QEE133 pKa = 3.95IVEE136 pKa = 4.08GRR138 pKa = 11.84VLNIGAIRR146 pKa = 11.84YY147 pKa = 7.51FEE149 pKa = 4.32RR150 pKa = 11.84LQSEE154 pKa = 4.57LDD156 pKa = 3.06SGYY159 pKa = 10.62QRR161 pKa = 11.84GHH163 pKa = 6.19QIAAYY168 pKa = 10.4LFGTPGTGKK177 pKa = 9.75SHH179 pKa = 6.88IAHH182 pKa = 6.78AAVEE186 pKa = 4.09EE187 pKa = 4.26RR188 pKa = 11.84TWYY191 pKa = 10.97SKK193 pKa = 11.18DD194 pKa = 3.1ATTKK198 pKa = 8.77WWDD201 pKa = 3.79GYY203 pKa = 10.84SGEE206 pKa = 4.36EE207 pKa = 4.13VVIVDD212 pKa = 4.01DD213 pKa = 3.94WSGPSTQAEE222 pKa = 3.75ADD224 pKa = 3.42QWKK227 pKa = 9.48KK228 pKa = 9.3WVDD231 pKa = 3.54RR232 pKa = 11.84YY233 pKa = 9.54PCRR236 pKa = 11.84VEE238 pKa = 4.08TKK240 pKa = 10.41GGHH243 pKa = 4.53VWLKK247 pKa = 7.19TKK249 pKa = 9.64QWIFTSNVRR258 pKa = 11.84WVALPVFFDD267 pKa = 3.36AALKK271 pKa = 10.55RR272 pKa = 11.84RR273 pKa = 11.84FIGNGG278 pKa = 3.03
Molecular weight: 31.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
553 |
275 |
278 |
276.5 |
31.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.787 ± 1.353 | 1.447 ± 0.269 |
5.063 ± 0.296 | 5.063 ± 2.182 |
4.702 ± 0.846 | 8.68 ± 1.341 |
2.893 ± 0.563 | 5.244 ± 0.667 |
4.34 ± 1.634 | 5.425 ± 0.298 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.085 ± 0.555 | 3.617 ± 1.116 |
4.34 ± 1.395 | 6.329 ± 1.265 |
6.329 ± 1.213 | 6.51 ± 0.302 |
7.595 ± 1.408 | 5.787 ± 0.802 |
3.797 ± 0.397 | 5.967 ± 1.815 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
