
Mesorhizobium sp. J18
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Mesorhizobium; unclassified Mesorhizobium
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
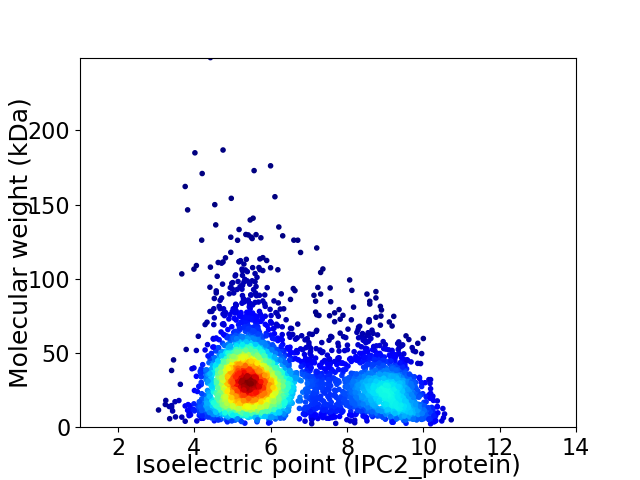
Virtual 2D-PAGE plot for 4171 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A562C1S7|A0A562C1S7_9RHIZ TetR family transcriptional regulator OS=Mesorhizobium sp. J18 OX=935263 GN=L598_000600001070 PE=4 SV=1
MM1 pKa = 7.03KK2 pKa = 9.7TVANARR8 pKa = 11.84GVNLYY13 pKa = 10.92YY14 pKa = 10.57SDD16 pKa = 3.34TSINHH21 pKa = 6.06FSATNSGPQLYY32 pKa = 7.54GTPDD36 pKa = 3.61NDD38 pKa = 4.44SLWGDD43 pKa = 3.62SSVDD47 pKa = 3.09VTMYY51 pKa = 10.91GGAGDD56 pKa = 5.07DD57 pKa = 3.1IYY59 pKa = 11.79YY60 pKa = 10.33LYY62 pKa = 11.2SPINRR67 pKa = 11.84AAEE70 pKa = 4.16DD71 pKa = 3.7PGEE74 pKa = 4.3GVDD77 pKa = 5.57TIVTWMSTRR86 pKa = 11.84LPDD89 pKa = 3.46NFEE92 pKa = 4.19NLTVTGSGRR101 pKa = 11.84YY102 pKa = 9.45AFGNAADD109 pKa = 4.48NIIKK113 pKa = 10.38GGSGQQTLDD122 pKa = 3.43GGSGNDD128 pKa = 3.37VLIGGGGADD137 pKa = 2.89IFVVSKK143 pKa = 11.4GNGSDD148 pKa = 4.54LIFDD152 pKa = 4.63FEE154 pKa = 4.39TTDD157 pKa = 3.22RR158 pKa = 11.84VRR160 pKa = 11.84LDD162 pKa = 3.47GYY164 pKa = 10.45GWNSFDD170 pKa = 4.33AVQDD174 pKa = 3.08RR175 pKa = 11.84MTQVGSDD182 pKa = 3.19VRR184 pKa = 11.84LDD186 pKa = 3.7LGVGEE191 pKa = 4.43ILVFKK196 pKa = 9.84DD197 pKa = 3.11TTIDD201 pKa = 3.54QFQADD206 pKa = 4.31QFEE209 pKa = 4.68LPLDD213 pKa = 4.11RR214 pKa = 11.84SAMSLSFAEE223 pKa = 4.46HH224 pKa = 6.76FNTLSLWDD232 pKa = 4.08GEE234 pKa = 4.41SGRR237 pKa = 11.84WDD239 pKa = 3.69SNFWWGAPNGSTLSGNSEE257 pKa = 4.2LQWYY261 pKa = 9.34IDD263 pKa = 3.61ANYY266 pKa = 10.53GPTSSVNPFSVEE278 pKa = 3.87NGVLTITAAPTPDD291 pKa = 3.9EE292 pKa = 4.61IKK294 pKa = 10.73PLINNYY300 pKa = 10.02EE301 pKa = 4.22YY302 pKa = 10.86TSGLLTTYY310 pKa = 10.59HH311 pKa = 6.53SFSQTYY317 pKa = 10.0GYY319 pKa = 11.18FEE321 pKa = 4.44MRR323 pKa = 11.84ADD325 pKa = 3.45MPEE328 pKa = 3.94NQGAWPAFWLLPADD342 pKa = 4.39GSWPPEE348 pKa = 3.55IDD350 pKa = 3.58VVEE353 pKa = 4.39MRR355 pKa = 11.84GQDD358 pKa = 3.36PNTIHH363 pKa = 5.66MTAHH367 pKa = 5.54TNEE370 pKa = 4.58TGSHH374 pKa = 4.81TMVSSAINVSDD385 pKa = 3.48TEE387 pKa = 4.23GFHH390 pKa = 6.52TYY392 pKa = 9.82GVLWTEE398 pKa = 5.5DD399 pKa = 3.37EE400 pKa = 4.44LVWYY404 pKa = 10.09FDD406 pKa = 3.89DD407 pKa = 3.91VAVARR412 pKa = 11.84AEE414 pKa = 4.49TPSDD418 pKa = 3.33MHH420 pKa = 8.33DD421 pKa = 3.43PMYY424 pKa = 10.44MLVNLAIGGPAGEE437 pKa = 5.01PDD439 pKa = 4.54DD440 pKa = 4.92GLAVPAEE447 pKa = 3.99MQIDD451 pKa = 4.22YY452 pKa = 10.79IRR454 pKa = 11.84AYY456 pKa = 9.92EE457 pKa = 4.08LDD459 pKa = 3.79EE460 pKa = 4.33VPEE463 pKa = 4.14VSLLANLFDD472 pKa = 4.02TLDD475 pKa = 3.37WLII478 pKa = 4.2
MM1 pKa = 7.03KK2 pKa = 9.7TVANARR8 pKa = 11.84GVNLYY13 pKa = 10.92YY14 pKa = 10.57SDD16 pKa = 3.34TSINHH21 pKa = 6.06FSATNSGPQLYY32 pKa = 7.54GTPDD36 pKa = 3.61NDD38 pKa = 4.44SLWGDD43 pKa = 3.62SSVDD47 pKa = 3.09VTMYY51 pKa = 10.91GGAGDD56 pKa = 5.07DD57 pKa = 3.1IYY59 pKa = 11.79YY60 pKa = 10.33LYY62 pKa = 11.2SPINRR67 pKa = 11.84AAEE70 pKa = 4.16DD71 pKa = 3.7PGEE74 pKa = 4.3GVDD77 pKa = 5.57TIVTWMSTRR86 pKa = 11.84LPDD89 pKa = 3.46NFEE92 pKa = 4.19NLTVTGSGRR101 pKa = 11.84YY102 pKa = 9.45AFGNAADD109 pKa = 4.48NIIKK113 pKa = 10.38GGSGQQTLDD122 pKa = 3.43GGSGNDD128 pKa = 3.37VLIGGGGADD137 pKa = 2.89IFVVSKK143 pKa = 11.4GNGSDD148 pKa = 4.54LIFDD152 pKa = 4.63FEE154 pKa = 4.39TTDD157 pKa = 3.22RR158 pKa = 11.84VRR160 pKa = 11.84LDD162 pKa = 3.47GYY164 pKa = 10.45GWNSFDD170 pKa = 4.33AVQDD174 pKa = 3.08RR175 pKa = 11.84MTQVGSDD182 pKa = 3.19VRR184 pKa = 11.84LDD186 pKa = 3.7LGVGEE191 pKa = 4.43ILVFKK196 pKa = 9.84DD197 pKa = 3.11TTIDD201 pKa = 3.54QFQADD206 pKa = 4.31QFEE209 pKa = 4.68LPLDD213 pKa = 4.11RR214 pKa = 11.84SAMSLSFAEE223 pKa = 4.46HH224 pKa = 6.76FNTLSLWDD232 pKa = 4.08GEE234 pKa = 4.41SGRR237 pKa = 11.84WDD239 pKa = 3.69SNFWWGAPNGSTLSGNSEE257 pKa = 4.2LQWYY261 pKa = 9.34IDD263 pKa = 3.61ANYY266 pKa = 10.53GPTSSVNPFSVEE278 pKa = 3.87NGVLTITAAPTPDD291 pKa = 3.9EE292 pKa = 4.61IKK294 pKa = 10.73PLINNYY300 pKa = 10.02EE301 pKa = 4.22YY302 pKa = 10.86TSGLLTTYY310 pKa = 10.59HH311 pKa = 6.53SFSQTYY317 pKa = 10.0GYY319 pKa = 11.18FEE321 pKa = 4.44MRR323 pKa = 11.84ADD325 pKa = 3.45MPEE328 pKa = 3.94NQGAWPAFWLLPADD342 pKa = 4.39GSWPPEE348 pKa = 3.55IDD350 pKa = 3.58VVEE353 pKa = 4.39MRR355 pKa = 11.84GQDD358 pKa = 3.36PNTIHH363 pKa = 5.66MTAHH367 pKa = 5.54TNEE370 pKa = 4.58TGSHH374 pKa = 4.81TMVSSAINVSDD385 pKa = 3.48TEE387 pKa = 4.23GFHH390 pKa = 6.52TYY392 pKa = 9.82GVLWTEE398 pKa = 5.5DD399 pKa = 3.37EE400 pKa = 4.44LVWYY404 pKa = 10.09FDD406 pKa = 3.89DD407 pKa = 3.91VAVARR412 pKa = 11.84AEE414 pKa = 4.49TPSDD418 pKa = 3.33MHH420 pKa = 8.33DD421 pKa = 3.43PMYY424 pKa = 10.44MLVNLAIGGPAGEE437 pKa = 5.01PDD439 pKa = 4.54DD440 pKa = 4.92GLAVPAEE447 pKa = 3.99MQIDD451 pKa = 4.22YY452 pKa = 10.79IRR454 pKa = 11.84AYY456 pKa = 9.92EE457 pKa = 4.08LDD459 pKa = 3.79EE460 pKa = 4.33VPEE463 pKa = 4.14VSLLANLFDD472 pKa = 4.02TLDD475 pKa = 3.37WLII478 pKa = 4.2
Molecular weight: 52.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A562CPY8|A0A562CPY8_9RHIZ AbrB family looped-hinge helix DNA binding protein OS=Mesorhizobium sp. J18 OX=935263 GN=L598_001500000800 PE=4 SV=1
MM1 pKa = 6.42TAGMKK6 pKa = 10.29RR7 pKa = 11.84EE8 pKa = 4.12DD9 pKa = 3.36RR10 pKa = 11.84SSRR13 pKa = 11.84EE14 pKa = 3.59KK15 pKa = 10.16MRR17 pKa = 11.84VSRR20 pKa = 11.84FLAAGPATIAVAGSGDD36 pKa = 3.67NLLLEE41 pKa = 4.44TEE43 pKa = 4.12NRR45 pKa = 11.84GTISVRR51 pKa = 11.84RR52 pKa = 11.84GEE54 pKa = 4.12FRR56 pKa = 11.84ALLAQGLVVVSGKK69 pKa = 9.12CARR72 pKa = 11.84LSAEE76 pKa = 3.91GQALARR82 pKa = 11.84RR83 pKa = 11.84AAATVEE89 pKa = 4.21PFRR92 pKa = 11.84AQHH95 pKa = 6.34GEE97 pKa = 3.93TGSVVMEE104 pKa = 3.94VQGMRR109 pKa = 11.84EE110 pKa = 3.83TVEE113 pKa = 3.92INLAEE118 pKa = 4.34SPLAQLASRR127 pKa = 11.84RR128 pKa = 11.84TRR130 pKa = 11.84KK131 pKa = 10.18GEE133 pKa = 3.83RR134 pKa = 11.84FLTGAEE140 pKa = 3.5IRR142 pKa = 11.84AGEE145 pKa = 4.41RR146 pKa = 11.84LRR148 pKa = 11.84ADD150 pKa = 3.46YY151 pKa = 10.1TRR153 pKa = 11.84GQIMPRR159 pKa = 11.84ITANWQAAVASGRR172 pKa = 11.84RR173 pKa = 11.84AGGSGGMVEE182 pKa = 4.5LTDD185 pKa = 3.45AALAARR191 pKa = 11.84QRR193 pKa = 11.84VDD195 pKa = 2.92RR196 pKa = 11.84ALQAVGPEE204 pKa = 4.07LAGVLVDD211 pKa = 3.39VCCFLKK217 pKa = 10.77GLEE220 pKa = 4.07RR221 pKa = 11.84VEE223 pKa = 4.49TEE225 pKa = 3.63RR226 pKa = 11.84GWPVRR231 pKa = 11.84SAKK234 pKa = 10.22VVLKK238 pKa = 9.34TALGALSRR246 pKa = 11.84HH247 pKa = 5.43YY248 pKa = 10.26EE249 pKa = 3.73PHH251 pKa = 6.53LCSGSQSRR259 pKa = 11.84PILHH263 pKa = 6.97WGAEE267 pKa = 4.56GYY269 pKa = 9.82RR270 pKa = 11.84PKK272 pKa = 10.84IMGG275 pKa = 3.69
MM1 pKa = 6.42TAGMKK6 pKa = 10.29RR7 pKa = 11.84EE8 pKa = 4.12DD9 pKa = 3.36RR10 pKa = 11.84SSRR13 pKa = 11.84EE14 pKa = 3.59KK15 pKa = 10.16MRR17 pKa = 11.84VSRR20 pKa = 11.84FLAAGPATIAVAGSGDD36 pKa = 3.67NLLLEE41 pKa = 4.44TEE43 pKa = 4.12NRR45 pKa = 11.84GTISVRR51 pKa = 11.84RR52 pKa = 11.84GEE54 pKa = 4.12FRR56 pKa = 11.84ALLAQGLVVVSGKK69 pKa = 9.12CARR72 pKa = 11.84LSAEE76 pKa = 3.91GQALARR82 pKa = 11.84RR83 pKa = 11.84AAATVEE89 pKa = 4.21PFRR92 pKa = 11.84AQHH95 pKa = 6.34GEE97 pKa = 3.93TGSVVMEE104 pKa = 3.94VQGMRR109 pKa = 11.84EE110 pKa = 3.83TVEE113 pKa = 3.92INLAEE118 pKa = 4.34SPLAQLASRR127 pKa = 11.84RR128 pKa = 11.84TRR130 pKa = 11.84KK131 pKa = 10.18GEE133 pKa = 3.83RR134 pKa = 11.84FLTGAEE140 pKa = 3.5IRR142 pKa = 11.84AGEE145 pKa = 4.41RR146 pKa = 11.84LRR148 pKa = 11.84ADD150 pKa = 3.46YY151 pKa = 10.1TRR153 pKa = 11.84GQIMPRR159 pKa = 11.84ITANWQAAVASGRR172 pKa = 11.84RR173 pKa = 11.84AGGSGGMVEE182 pKa = 4.5LTDD185 pKa = 3.45AALAARR191 pKa = 11.84QRR193 pKa = 11.84VDD195 pKa = 2.92RR196 pKa = 11.84ALQAVGPEE204 pKa = 4.07LAGVLVDD211 pKa = 3.39VCCFLKK217 pKa = 10.77GLEE220 pKa = 4.07RR221 pKa = 11.84VEE223 pKa = 4.49TEE225 pKa = 3.63RR226 pKa = 11.84GWPVRR231 pKa = 11.84SAKK234 pKa = 10.22VVLKK238 pKa = 9.34TALGALSRR246 pKa = 11.84HH247 pKa = 5.43YY248 pKa = 10.26EE249 pKa = 3.73PHH251 pKa = 6.53LCSGSQSRR259 pKa = 11.84PILHH263 pKa = 6.97WGAEE267 pKa = 4.56GYY269 pKa = 9.82RR270 pKa = 11.84PKK272 pKa = 10.84IMGG275 pKa = 3.69
Molecular weight: 29.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1255211 |
22 |
2297 |
300.9 |
32.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.801 ± 0.045 | 0.811 ± 0.013 |
5.556 ± 0.034 | 6.456 ± 0.039 |
3.874 ± 0.026 | 8.604 ± 0.041 |
2.011 ± 0.02 | 5.629 ± 0.028 |
3.433 ± 0.029 | 9.774 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.616 ± 0.017 | 2.657 ± 0.021 |
5.041 ± 0.026 | 2.935 ± 0.021 |
7.253 ± 0.036 | 5.459 ± 0.027 |
5.135 ± 0.026 | 7.339 ± 0.03 |
1.294 ± 0.015 | 2.307 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
