
Beihai sea slater virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
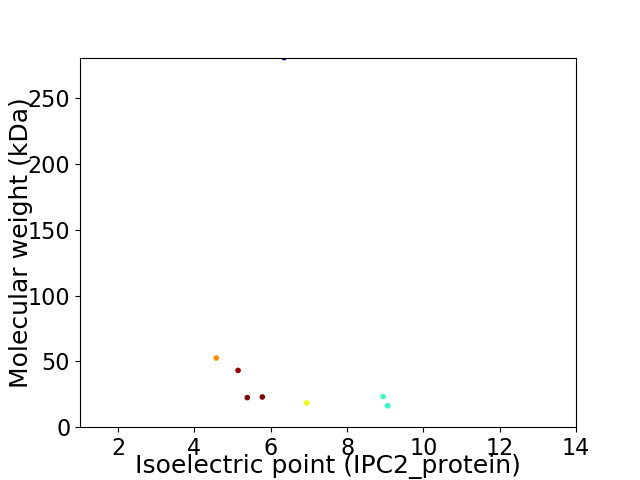
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
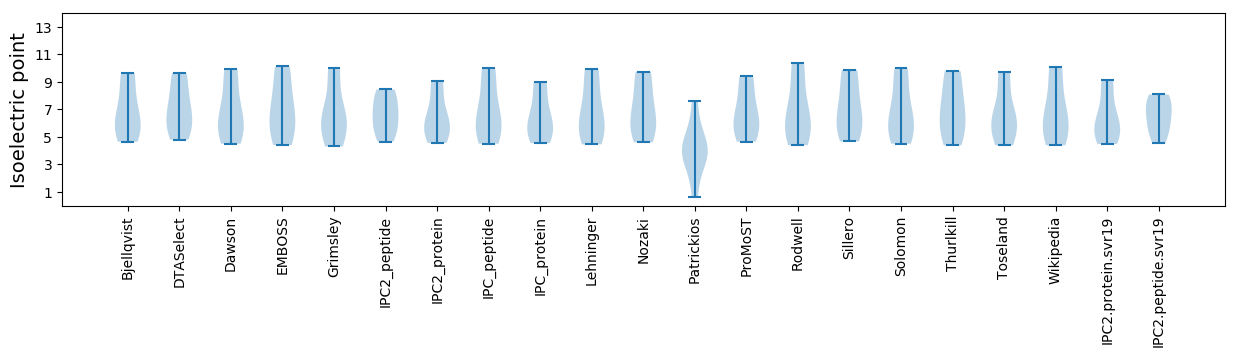
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJK8|A0A1L3KJK8_9VIRU RdRp OS=Beihai sea slater virus 4 OX=1922660 PE=4 SV=1
MM1 pKa = 6.89NRR3 pKa = 11.84NYY5 pKa = 10.0FRR7 pKa = 11.84LVHH10 pKa = 5.85HH11 pKa = 6.36QFNEE15 pKa = 3.86LRR17 pKa = 11.84RR18 pKa = 11.84DD19 pKa = 3.6TQVAYY24 pKa = 10.45ASLAALLLVAGLATTGIFTLQEE46 pKa = 3.39ISEE49 pKa = 4.66KK50 pKa = 10.12IEE52 pKa = 4.12HH53 pKa = 6.85DD54 pKa = 3.89PEE56 pKa = 3.87HH57 pKa = 6.53TIIDD61 pKa = 3.99GVIDD65 pKa = 4.73DD66 pKa = 4.74VIEE69 pKa = 4.19MYY71 pKa = 10.97LNQLFTQPGLASRR84 pKa = 11.84ITALGDD90 pKa = 3.78LIKK93 pKa = 10.53SIKK96 pKa = 9.89EE97 pKa = 3.58IIGPILKK104 pKa = 10.31YY105 pKa = 8.9FTKK108 pKa = 10.74DD109 pKa = 2.62SDD111 pKa = 4.05YY112 pKa = 11.75LLGNVTAILTNITYY126 pKa = 10.86SNLVSDD132 pKa = 6.37FILQEE137 pKa = 4.81LIDD140 pKa = 4.75LNCAIHH146 pKa = 6.3KK147 pKa = 10.22CEE149 pKa = 5.77SIDD152 pKa = 3.72EE153 pKa = 4.78DD154 pKa = 5.07DD155 pKa = 4.33VSDD158 pKa = 3.99MLNSILDD165 pKa = 4.23FIEE168 pKa = 4.3DD169 pKa = 3.77HH170 pKa = 6.76PALPEE175 pKa = 3.73KK176 pKa = 10.8VYY178 pKa = 10.77DD179 pKa = 3.82YY180 pKa = 11.12FKK182 pKa = 10.11TYY184 pKa = 10.22PITAEE189 pKa = 3.87CLHH192 pKa = 6.78FKK194 pKa = 10.63RR195 pKa = 11.84IALLSDD201 pKa = 4.31ADD203 pKa = 4.11TQQQLSARR211 pKa = 11.84PDD213 pKa = 3.17HH214 pKa = 6.28EE215 pKa = 5.15AYY217 pKa = 10.0EE218 pKa = 4.57VVRR221 pKa = 11.84HH222 pKa = 5.49CVSNGVNCQPVRR234 pKa = 11.84YY235 pKa = 9.02EE236 pKa = 4.47SKK238 pKa = 10.97CSDD241 pKa = 2.67GKK243 pKa = 10.77YY244 pKa = 9.91YY245 pKa = 11.04CFDD248 pKa = 4.48DD249 pKa = 4.59GFFAIPTYY257 pKa = 10.71EE258 pKa = 3.93PFSFKK263 pKa = 10.67VLQNEE268 pKa = 4.11DD269 pKa = 3.32WVMEE273 pKa = 4.31GADD276 pKa = 3.94YY277 pKa = 11.08TDD279 pKa = 3.69LVYY282 pKa = 10.45FQGKK286 pKa = 8.38IRR288 pKa = 11.84YY289 pKa = 6.34TPQAYY294 pKa = 8.11TYY296 pKa = 9.85KK297 pKa = 10.28FQYY300 pKa = 9.6LKK302 pKa = 11.02CVDD305 pKa = 4.27GEE307 pKa = 4.17LHH309 pKa = 7.32IYY311 pKa = 8.38TCEE314 pKa = 3.97DD315 pKa = 3.33TVEE318 pKa = 4.1SCEE321 pKa = 4.37LYY323 pKa = 10.9NLEE326 pKa = 4.23TTCYY330 pKa = 10.96GEE332 pKa = 4.04GKK334 pKa = 10.5YY335 pKa = 10.51LFLPMHH341 pKa = 6.59VKK343 pKa = 10.51RR344 pKa = 11.84LDD346 pKa = 3.71EE347 pKa = 4.33TWCSIMPDD355 pKa = 3.34QAEE358 pKa = 4.23SCSIGDD364 pKa = 3.4SWVDD368 pKa = 2.99SFYY371 pKa = 11.09YY372 pKa = 10.33DD373 pKa = 2.94FWYY376 pKa = 10.1FIVNGAFGTHH386 pKa = 4.62VTAEE390 pKa = 4.34FKK392 pKa = 10.82DD393 pKa = 3.64QEE395 pKa = 4.58SFTKK399 pKa = 10.53KK400 pKa = 8.31STAYY404 pKa = 8.79TYY406 pKa = 10.08CHH408 pKa = 6.72KK409 pKa = 10.27CDD411 pKa = 3.57EE412 pKa = 4.66GPYY415 pKa = 9.71PSGSEE420 pKa = 4.26LLIAMDD426 pKa = 3.81NFVNASFPTQEE437 pKa = 3.75STYY440 pKa = 11.24GGMGNGKK447 pKa = 8.56HH448 pKa = 6.19LCLNKK453 pKa = 10.49YY454 pKa = 10.38NLFKK458 pKa = 11.12
MM1 pKa = 6.89NRR3 pKa = 11.84NYY5 pKa = 10.0FRR7 pKa = 11.84LVHH10 pKa = 5.85HH11 pKa = 6.36QFNEE15 pKa = 3.86LRR17 pKa = 11.84RR18 pKa = 11.84DD19 pKa = 3.6TQVAYY24 pKa = 10.45ASLAALLLVAGLATTGIFTLQEE46 pKa = 3.39ISEE49 pKa = 4.66KK50 pKa = 10.12IEE52 pKa = 4.12HH53 pKa = 6.85DD54 pKa = 3.89PEE56 pKa = 3.87HH57 pKa = 6.53TIIDD61 pKa = 3.99GVIDD65 pKa = 4.73DD66 pKa = 4.74VIEE69 pKa = 4.19MYY71 pKa = 10.97LNQLFTQPGLASRR84 pKa = 11.84ITALGDD90 pKa = 3.78LIKK93 pKa = 10.53SIKK96 pKa = 9.89EE97 pKa = 3.58IIGPILKK104 pKa = 10.31YY105 pKa = 8.9FTKK108 pKa = 10.74DD109 pKa = 2.62SDD111 pKa = 4.05YY112 pKa = 11.75LLGNVTAILTNITYY126 pKa = 10.86SNLVSDD132 pKa = 6.37FILQEE137 pKa = 4.81LIDD140 pKa = 4.75LNCAIHH146 pKa = 6.3KK147 pKa = 10.22CEE149 pKa = 5.77SIDD152 pKa = 3.72EE153 pKa = 4.78DD154 pKa = 5.07DD155 pKa = 4.33VSDD158 pKa = 3.99MLNSILDD165 pKa = 4.23FIEE168 pKa = 4.3DD169 pKa = 3.77HH170 pKa = 6.76PALPEE175 pKa = 3.73KK176 pKa = 10.8VYY178 pKa = 10.77DD179 pKa = 3.82YY180 pKa = 11.12FKK182 pKa = 10.11TYY184 pKa = 10.22PITAEE189 pKa = 3.87CLHH192 pKa = 6.78FKK194 pKa = 10.63RR195 pKa = 11.84IALLSDD201 pKa = 4.31ADD203 pKa = 4.11TQQQLSARR211 pKa = 11.84PDD213 pKa = 3.17HH214 pKa = 6.28EE215 pKa = 5.15AYY217 pKa = 10.0EE218 pKa = 4.57VVRR221 pKa = 11.84HH222 pKa = 5.49CVSNGVNCQPVRR234 pKa = 11.84YY235 pKa = 9.02EE236 pKa = 4.47SKK238 pKa = 10.97CSDD241 pKa = 2.67GKK243 pKa = 10.77YY244 pKa = 9.91YY245 pKa = 11.04CFDD248 pKa = 4.48DD249 pKa = 4.59GFFAIPTYY257 pKa = 10.71EE258 pKa = 3.93PFSFKK263 pKa = 10.67VLQNEE268 pKa = 4.11DD269 pKa = 3.32WVMEE273 pKa = 4.31GADD276 pKa = 3.94YY277 pKa = 11.08TDD279 pKa = 3.69LVYY282 pKa = 10.45FQGKK286 pKa = 8.38IRR288 pKa = 11.84YY289 pKa = 6.34TPQAYY294 pKa = 8.11TYY296 pKa = 9.85KK297 pKa = 10.28FQYY300 pKa = 9.6LKK302 pKa = 11.02CVDD305 pKa = 4.27GEE307 pKa = 4.17LHH309 pKa = 7.32IYY311 pKa = 8.38TCEE314 pKa = 3.97DD315 pKa = 3.33TVEE318 pKa = 4.1SCEE321 pKa = 4.37LYY323 pKa = 10.9NLEE326 pKa = 4.23TTCYY330 pKa = 10.96GEE332 pKa = 4.04GKK334 pKa = 10.5YY335 pKa = 10.51LFLPMHH341 pKa = 6.59VKK343 pKa = 10.51RR344 pKa = 11.84LDD346 pKa = 3.71EE347 pKa = 4.33TWCSIMPDD355 pKa = 3.34QAEE358 pKa = 4.23SCSIGDD364 pKa = 3.4SWVDD368 pKa = 2.99SFYY371 pKa = 11.09YY372 pKa = 10.33DD373 pKa = 2.94FWYY376 pKa = 10.1FIVNGAFGTHH386 pKa = 4.62VTAEE390 pKa = 4.34FKK392 pKa = 10.82DD393 pKa = 3.64QEE395 pKa = 4.58SFTKK399 pKa = 10.53KK400 pKa = 8.31STAYY404 pKa = 8.79TYY406 pKa = 10.08CHH408 pKa = 6.72KK409 pKa = 10.27CDD411 pKa = 3.57EE412 pKa = 4.66GPYY415 pKa = 9.71PSGSEE420 pKa = 4.26LLIAMDD426 pKa = 3.81NFVNASFPTQEE437 pKa = 3.75STYY440 pKa = 11.24GGMGNGKK447 pKa = 8.56HH448 pKa = 6.19LCLNKK453 pKa = 10.49YY454 pKa = 10.38NLFKK458 pKa = 11.12
Molecular weight: 52.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJP7|A0A1L3KJP7_9VIRU Uncharacterized protein OS=Beihai sea slater virus 4 OX=1922660 PE=4 SV=1
MM1 pKa = 7.93IMAKK5 pKa = 10.42GDD7 pKa = 3.8DD8 pKa = 4.19LYY10 pKa = 11.05IHH12 pKa = 7.18AKK14 pKa = 10.28KK15 pKa = 10.32IIQKK19 pKa = 10.21HH20 pKa = 4.09FDD22 pKa = 3.39KK23 pKa = 10.76NFKK26 pKa = 9.52ISRR29 pKa = 11.84PIVPSFVGFIIYY41 pKa = 10.07DD42 pKa = 3.74KK43 pKa = 10.99LYY45 pKa = 11.11LDD47 pKa = 3.77IPKK50 pKa = 9.17LTVRR54 pKa = 11.84VLNRR58 pKa = 11.84NYY60 pKa = 10.97ADD62 pKa = 3.5NKK64 pKa = 10.49SISDD68 pKa = 3.72YY69 pKa = 10.91IIAVKK74 pKa = 10.42DD75 pKa = 3.23WLSVIKK81 pKa = 10.32SQEE84 pKa = 3.89HH85 pKa = 5.08LTVCSILNAKK95 pKa = 10.21RR96 pKa = 11.84YY97 pKa = 8.07GLSVAEE103 pKa = 4.51CEE105 pKa = 4.28YY106 pKa = 11.57LLGFLFAFSRR116 pKa = 11.84GRR118 pKa = 11.84IINTTNSKK126 pKa = 10.06ILVRR130 pKa = 11.84EE131 pKa = 4.16RR132 pKa = 11.84KK133 pKa = 9.27QPFVLNEE140 pKa = 3.7
MM1 pKa = 7.93IMAKK5 pKa = 10.42GDD7 pKa = 3.8DD8 pKa = 4.19LYY10 pKa = 11.05IHH12 pKa = 7.18AKK14 pKa = 10.28KK15 pKa = 10.32IIQKK19 pKa = 10.21HH20 pKa = 4.09FDD22 pKa = 3.39KK23 pKa = 10.76NFKK26 pKa = 9.52ISRR29 pKa = 11.84PIVPSFVGFIIYY41 pKa = 10.07DD42 pKa = 3.74KK43 pKa = 10.99LYY45 pKa = 11.11LDD47 pKa = 3.77IPKK50 pKa = 9.17LTVRR54 pKa = 11.84VLNRR58 pKa = 11.84NYY60 pKa = 10.97ADD62 pKa = 3.5NKK64 pKa = 10.49SISDD68 pKa = 3.72YY69 pKa = 10.91IIAVKK74 pKa = 10.42DD75 pKa = 3.23WLSVIKK81 pKa = 10.32SQEE84 pKa = 3.89HH85 pKa = 5.08LTVCSILNAKK95 pKa = 10.21RR96 pKa = 11.84YY97 pKa = 8.07GLSVAEE103 pKa = 4.51CEE105 pKa = 4.28YY106 pKa = 11.57LLGFLFAFSRR116 pKa = 11.84GRR118 pKa = 11.84IINTTNSKK126 pKa = 10.06ILVRR130 pKa = 11.84EE131 pKa = 4.16RR132 pKa = 11.84KK133 pKa = 9.27QPFVLNEE140 pKa = 3.7
Molecular weight: 16.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4210 |
140 |
2462 |
526.3 |
59.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.081 ± 0.545 | 2.09 ± 0.266 |
6.722 ± 0.635 | 6.01 ± 0.764 |
4.679 ± 0.431 | 4.537 ± 0.358 |
2.779 ± 0.272 | 6.318 ± 0.659 |
7.007 ± 1.385 | 8.622 ± 0.837 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.09 ± 0.27 | 4.751 ± 0.291 |
4.276 ± 0.332 | 3.492 ± 0.256 |
4.751 ± 0.497 | 6.675 ± 0.788 |
6.698 ± 0.476 | 7.031 ± 0.402 |
0.998 ± 0.163 | 4.394 ± 0.649 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
