
Treponema brennaborense (strain DSM 12168 / CIP 105900 / DD5/3)
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Treponemataceae; Treponema; Treponema brennaborense
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
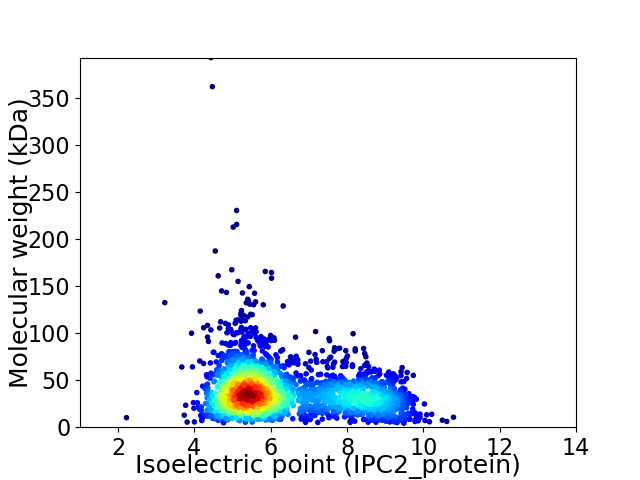
Virtual 2D-PAGE plot for 2522 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
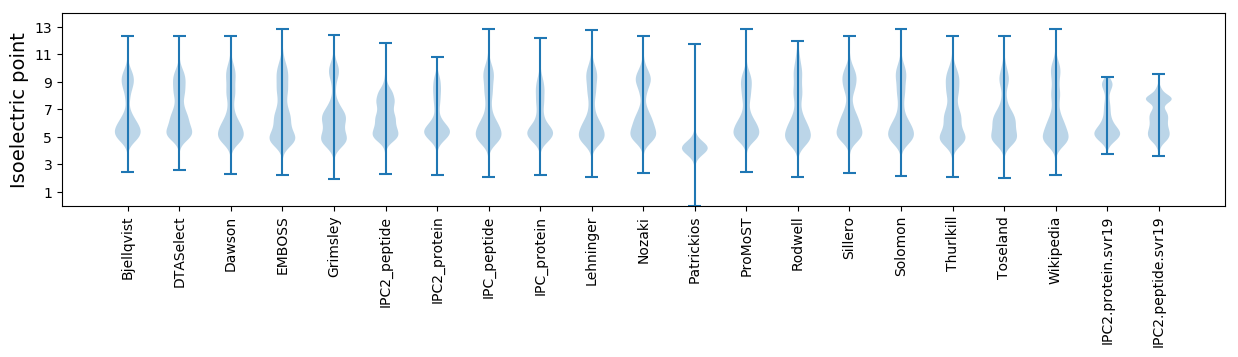
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F4LML1|F4LML1_TREBD Uncharacterized protein OS=Treponema brennaborense (strain DSM 12168 / CIP 105900 / DD5/3) OX=906968 GN=Trebr_1333 PE=4 SV=1
MM1 pKa = 7.19YY2 pKa = 10.82VNNDD6 pKa = 3.22PVNYY10 pKa = 9.59VDD12 pKa = 5.39LWGLSAGDD20 pKa = 4.59AKK22 pKa = 10.87SVWDD26 pKa = 4.08GFTGAGNPYY35 pKa = 10.54ANYY38 pKa = 10.06GVSTIIDD45 pKa = 3.73STLSSDD51 pKa = 3.55IAEE54 pKa = 4.22KK55 pKa = 7.46TTNYY59 pKa = 10.19VNDD62 pKa = 4.46FRR64 pKa = 11.84CDD66 pKa = 3.09MYY68 pKa = 11.29AWNSAVSDD76 pKa = 3.76GVNPKK81 pKa = 9.52GQNGEE86 pKa = 3.88DD87 pKa = 3.27WDD89 pKa = 3.73IDD91 pKa = 3.79RR92 pKa = 11.84EE93 pKa = 4.58SVSDD97 pKa = 3.86IYY99 pKa = 11.78NKK101 pKa = 10.16FPVLRR106 pKa = 11.84FVAPQNNTKK115 pKa = 10.37GFAFYY120 pKa = 10.55FNDD123 pKa = 3.57MNDD126 pKa = 3.53EE127 pKa = 4.25PVHH130 pKa = 7.51IEE132 pKa = 4.22YY133 pKa = 10.93YY134 pKa = 11.0DD135 pKa = 3.49NTNDD139 pKa = 3.19VSEE142 pKa = 4.85TYY144 pKa = 10.39VQYY147 pKa = 9.4KK148 pKa = 9.55TDD150 pKa = 3.25GKK152 pKa = 10.48NAVEE156 pKa = 4.17TVDD159 pKa = 5.15RR160 pKa = 11.84KK161 pKa = 10.77INDD164 pKa = 3.58DD165 pKa = 3.74RR166 pKa = 11.84NLSVVYY172 pKa = 9.12VVIYY176 pKa = 10.9SEE178 pKa = 4.14
MM1 pKa = 7.19YY2 pKa = 10.82VNNDD6 pKa = 3.22PVNYY10 pKa = 9.59VDD12 pKa = 5.39LWGLSAGDD20 pKa = 4.59AKK22 pKa = 10.87SVWDD26 pKa = 4.08GFTGAGNPYY35 pKa = 10.54ANYY38 pKa = 10.06GVSTIIDD45 pKa = 3.73STLSSDD51 pKa = 3.55IAEE54 pKa = 4.22KK55 pKa = 7.46TTNYY59 pKa = 10.19VNDD62 pKa = 4.46FRR64 pKa = 11.84CDD66 pKa = 3.09MYY68 pKa = 11.29AWNSAVSDD76 pKa = 3.76GVNPKK81 pKa = 9.52GQNGEE86 pKa = 3.88DD87 pKa = 3.27WDD89 pKa = 3.73IDD91 pKa = 3.79RR92 pKa = 11.84EE93 pKa = 4.58SVSDD97 pKa = 3.86IYY99 pKa = 11.78NKK101 pKa = 10.16FPVLRR106 pKa = 11.84FVAPQNNTKK115 pKa = 10.37GFAFYY120 pKa = 10.55FNDD123 pKa = 3.57MNDD126 pKa = 3.53EE127 pKa = 4.25PVHH130 pKa = 7.51IEE132 pKa = 4.22YY133 pKa = 10.93YY134 pKa = 11.0DD135 pKa = 3.49NTNDD139 pKa = 3.19VSEE142 pKa = 4.85TYY144 pKa = 10.39VQYY147 pKa = 9.4KK148 pKa = 9.55TDD150 pKa = 3.25GKK152 pKa = 10.48NAVEE156 pKa = 4.17TVDD159 pKa = 5.15RR160 pKa = 11.84KK161 pKa = 10.77INDD164 pKa = 3.58DD165 pKa = 3.74RR166 pKa = 11.84NLSVVYY172 pKa = 9.12VVIYY176 pKa = 10.9SEE178 pKa = 4.14
Molecular weight: 20.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F4LNE6|F4LNE6_TREBD Putative signal transduction protein OS=Treponema brennaborense (strain DSM 12168 / CIP 105900 / DD5/3) OX=906968 GN=Trebr_2498 PE=4 SV=1
MM1 pKa = 7.57KK2 pKa = 10.03ISAPHH7 pKa = 6.1PVRR10 pKa = 11.84TGTSLTVRR18 pKa = 11.84FTATALFLLAAAAAGIAFGTEE39 pKa = 4.06RR40 pKa = 11.84IPFRR44 pKa = 11.84EE45 pKa = 3.73IGNALFRR52 pKa = 11.84PAAADD57 pKa = 3.3PFAAVIVRR65 pKa = 11.84EE66 pKa = 3.94LRR68 pKa = 11.84LPRR71 pKa = 11.84VLLAGLSGALLASAGAAFQGFFRR94 pKa = 11.84NPLAEE99 pKa = 4.21SGIMGISSGAALGAVLSAFIPAATGIAGAAEE130 pKa = 4.03LASRR134 pKa = 11.84FPQFMQANVTTVCAFCGAAAAAFLIYY160 pKa = 10.25AASKK164 pKa = 8.7LTARR168 pKa = 11.84SGGTIAILLTGTAAGTFFSAVTSVVLLVKK197 pKa = 9.89DD198 pKa = 4.05TEE200 pKa = 4.21LHH202 pKa = 7.06RR203 pKa = 11.84MFVWTLGSFNGKK215 pKa = 8.56GWSEE219 pKa = 3.94LAFVLPAAVLSAVLLAGCARR239 pKa = 11.84FLDD242 pKa = 3.96VLSGGEE248 pKa = 3.9RR249 pKa = 11.84TAQALGLDD257 pKa = 4.05PAHH260 pKa = 6.93ARR262 pKa = 11.84LLVLTAGSLASAAAVCAGGTIGFVGLIAPHH292 pKa = 4.99VARR295 pKa = 11.84RR296 pKa = 11.84LFSPRR301 pKa = 11.84HH302 pKa = 5.15AVLIPASMMGGAVFLITCDD321 pKa = 3.1TAARR325 pKa = 11.84TVAAPAEE332 pKa = 4.23IPVGIITALAGAPFFISILFGGRR355 pKa = 11.84NRR357 pKa = 11.84TGNFNEE363 pKa = 4.18
MM1 pKa = 7.57KK2 pKa = 10.03ISAPHH7 pKa = 6.1PVRR10 pKa = 11.84TGTSLTVRR18 pKa = 11.84FTATALFLLAAAAAGIAFGTEE39 pKa = 4.06RR40 pKa = 11.84IPFRR44 pKa = 11.84EE45 pKa = 3.73IGNALFRR52 pKa = 11.84PAAADD57 pKa = 3.3PFAAVIVRR65 pKa = 11.84EE66 pKa = 3.94LRR68 pKa = 11.84LPRR71 pKa = 11.84VLLAGLSGALLASAGAAFQGFFRR94 pKa = 11.84NPLAEE99 pKa = 4.21SGIMGISSGAALGAVLSAFIPAATGIAGAAEE130 pKa = 4.03LASRR134 pKa = 11.84FPQFMQANVTTVCAFCGAAAAAFLIYY160 pKa = 10.25AASKK164 pKa = 8.7LTARR168 pKa = 11.84SGGTIAILLTGTAAGTFFSAVTSVVLLVKK197 pKa = 9.89DD198 pKa = 4.05TEE200 pKa = 4.21LHH202 pKa = 7.06RR203 pKa = 11.84MFVWTLGSFNGKK215 pKa = 8.56GWSEE219 pKa = 3.94LAFVLPAAVLSAVLLAGCARR239 pKa = 11.84FLDD242 pKa = 3.96VLSGGEE248 pKa = 3.9RR249 pKa = 11.84TAQALGLDD257 pKa = 4.05PAHH260 pKa = 6.93ARR262 pKa = 11.84LLVLTAGSLASAAAVCAGGTIGFVGLIAPHH292 pKa = 4.99VARR295 pKa = 11.84RR296 pKa = 11.84LFSPRR301 pKa = 11.84HH302 pKa = 5.15AVLIPASMMGGAVFLITCDD321 pKa = 3.1TAARR325 pKa = 11.84TVAAPAEE332 pKa = 4.23IPVGIITALAGAPFFISILFGGRR355 pKa = 11.84NRR357 pKa = 11.84TGNFNEE363 pKa = 4.18
Molecular weight: 36.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
908311 |
37 |
3722 |
360.2 |
39.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.076 ± 0.084 | 1.456 ± 0.018 |
5.441 ± 0.037 | 6.136 ± 0.058 |
4.719 ± 0.04 | 7.16 ± 0.048 |
1.48 ± 0.019 | 6.39 ± 0.046 |
5.406 ± 0.045 | 9.214 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.255 ± 0.025 | 3.807 ± 0.039 |
4.08 ± 0.036 | 2.955 ± 0.024 |
5.17 ± 0.046 | 6.677 ± 0.042 |
5.884 ± 0.046 | 7.152 ± 0.045 |
0.948 ± 0.014 | 3.593 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
