
Porcine hemagglutinating encephalomyelitis virus (strain 67N) (HEV-67N)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Embecovirus; Betacoronavirus 1; Porcine hemagglutinating encephalomyelitis virus
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
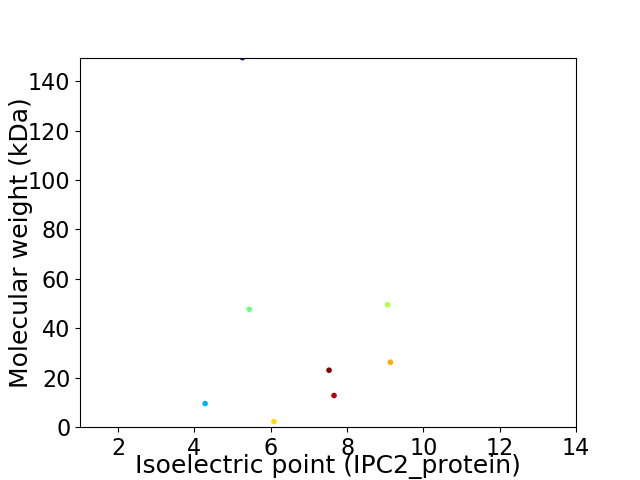
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q774I1|NS12_CVP67 Non-structural protein of 12.7 kDa OS=Porcine hemagglutinating encephalomyelitis virus (strain 67N) OX=230237 PE=3 SV=1
MM1 pKa = 7.17FMADD5 pKa = 4.32AYY7 pKa = 11.22LADD10 pKa = 3.99TVWYY14 pKa = 8.76VGQIIFIVAICLLVIIVVVAFLATFKK40 pKa = 11.14LCIQLCGMCNTLVLSPSIYY59 pKa = 9.46VFNRR63 pKa = 11.84GRR65 pKa = 11.84QFYY68 pKa = 10.64EE69 pKa = 4.42FYY71 pKa = 11.08NDD73 pKa = 3.24VKK75 pKa = 11.05PPVLDD80 pKa = 3.51VDD82 pKa = 4.53DD83 pKa = 4.05VV84 pKa = 3.68
MM1 pKa = 7.17FMADD5 pKa = 4.32AYY7 pKa = 11.22LADD10 pKa = 3.99TVWYY14 pKa = 8.76VGQIIFIVAICLLVIIVVVAFLATFKK40 pKa = 11.14LCIQLCGMCNTLVLSPSIYY59 pKa = 9.46VFNRR63 pKa = 11.84GRR65 pKa = 11.84QFYY68 pKa = 10.64EE69 pKa = 4.42FYY71 pKa = 11.08NDD73 pKa = 3.24VKK75 pKa = 11.05PPVLDD80 pKa = 3.51VDD82 pKa = 4.53DD83 pKa = 4.05VV84 pKa = 3.68
Molecular weight: 9.55 kDa
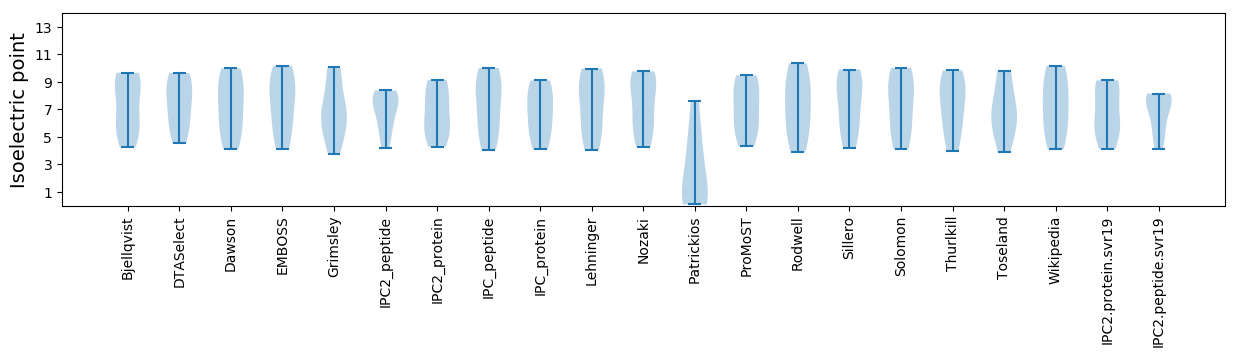
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q8BB24|VME1_CVP67 Membrane protein OS=Porcine hemagglutinating encephalomyelitis virus (strain 67N) OX=230237 GN=M PE=3 SV=1
MM1 pKa = 7.55SFTPGKK7 pKa = 9.79QSSSRR12 pKa = 11.84ASSGNRR18 pKa = 11.84SGNGILKK25 pKa = 9.42WADD28 pKa = 3.13QSDD31 pKa = 3.63QSRR34 pKa = 11.84NVQTRR39 pKa = 11.84GRR41 pKa = 11.84RR42 pKa = 11.84VQSKK46 pKa = 7.32QTATSQQPSGGTVVPYY62 pKa = 9.51YY63 pKa = 10.8SWFSGITQFQKK74 pKa = 10.77GKK76 pKa = 9.42EE77 pKa = 3.95FEE79 pKa = 4.16FAEE82 pKa = 4.69GQGVPIAPGVPSTEE96 pKa = 4.06AKK98 pKa = 10.31GYY100 pKa = 8.63WYY102 pKa = 10.34RR103 pKa = 11.84HH104 pKa = 4.43NRR106 pKa = 11.84RR107 pKa = 11.84SFKK110 pKa = 10.37TADD113 pKa = 3.13GNQRR117 pKa = 11.84QLLPRR122 pKa = 11.84WYY124 pKa = 9.8FYY126 pKa = 11.56YY127 pKa = 10.78LGTGPHH133 pKa = 6.97AKK135 pKa = 9.91DD136 pKa = 3.92QYY138 pKa = 10.35GTDD141 pKa = 3.18IDD143 pKa = 4.14GVFWVASNQADD154 pKa = 3.42INTPADD160 pKa = 3.29IVDD163 pKa = 4.43RR164 pKa = 11.84DD165 pKa = 3.85PSSDD169 pKa = 3.16EE170 pKa = 4.8AIPTRR175 pKa = 11.84FPPGTVLPQGYY186 pKa = 9.38YY187 pKa = 9.56IEE189 pKa = 4.86GSGRR193 pKa = 11.84SAPNSRR199 pKa = 11.84STSRR203 pKa = 11.84APNRR207 pKa = 11.84APSAGSRR214 pKa = 11.84SRR216 pKa = 11.84ANSGNRR222 pKa = 11.84TSTPGVTPDD231 pKa = 3.56MADD234 pKa = 3.28QIASLVLAKK243 pKa = 10.41LGKK246 pKa = 10.07DD247 pKa = 2.95ATKK250 pKa = 9.49PQQVTKK256 pKa = 9.36QTAKK260 pKa = 10.33EE261 pKa = 4.03VRR263 pKa = 11.84QKK265 pKa = 10.72ILNKK269 pKa = 9.23PRR271 pKa = 11.84QKK273 pKa = 10.46RR274 pKa = 11.84SPNKK278 pKa = 9.02QCTVQQCFGKK288 pKa = 10.48RR289 pKa = 11.84GPNQNFGGGEE299 pKa = 3.99MLKK302 pKa = 10.67LGTSDD307 pKa = 3.38PQFPILAEE315 pKa = 4.05LAPTAGAFFFGSRR328 pKa = 11.84LEE330 pKa = 4.03LAKK333 pKa = 10.7VQNLSGNPDD342 pKa = 3.48EE343 pKa = 4.7PQKK346 pKa = 11.04DD347 pKa = 4.22VYY349 pKa = 9.8EE350 pKa = 3.97LRR352 pKa = 11.84YY353 pKa = 10.38NGAIRR358 pKa = 11.84FDD360 pKa = 3.73STLSGFEE367 pKa = 4.32TIMKK371 pKa = 9.76VLNQNLNAYY380 pKa = 7.22QHH382 pKa = 6.17QEE384 pKa = 3.85DD385 pKa = 3.9GMMNISPKK393 pKa = 8.82PQRR396 pKa = 11.84QRR398 pKa = 11.84GQKK401 pKa = 9.61NGQVEE406 pKa = 4.3NDD408 pKa = 3.75NVSVAAPKK416 pKa = 10.71SRR418 pKa = 11.84VQQNKK423 pKa = 9.07SRR425 pKa = 11.84EE426 pKa = 4.15LTAEE430 pKa = 4.92DD431 pKa = 3.26ISLLKK436 pKa = 11.01KK437 pKa = 9.61MDD439 pKa = 3.6EE440 pKa = 4.96PYY442 pKa = 10.5TEE444 pKa = 4.35DD445 pKa = 3.07TSEE448 pKa = 3.68II449 pKa = 3.91
MM1 pKa = 7.55SFTPGKK7 pKa = 9.79QSSSRR12 pKa = 11.84ASSGNRR18 pKa = 11.84SGNGILKK25 pKa = 9.42WADD28 pKa = 3.13QSDD31 pKa = 3.63QSRR34 pKa = 11.84NVQTRR39 pKa = 11.84GRR41 pKa = 11.84RR42 pKa = 11.84VQSKK46 pKa = 7.32QTATSQQPSGGTVVPYY62 pKa = 9.51YY63 pKa = 10.8SWFSGITQFQKK74 pKa = 10.77GKK76 pKa = 9.42EE77 pKa = 3.95FEE79 pKa = 4.16FAEE82 pKa = 4.69GQGVPIAPGVPSTEE96 pKa = 4.06AKK98 pKa = 10.31GYY100 pKa = 8.63WYY102 pKa = 10.34RR103 pKa = 11.84HH104 pKa = 4.43NRR106 pKa = 11.84RR107 pKa = 11.84SFKK110 pKa = 10.37TADD113 pKa = 3.13GNQRR117 pKa = 11.84QLLPRR122 pKa = 11.84WYY124 pKa = 9.8FYY126 pKa = 11.56YY127 pKa = 10.78LGTGPHH133 pKa = 6.97AKK135 pKa = 9.91DD136 pKa = 3.92QYY138 pKa = 10.35GTDD141 pKa = 3.18IDD143 pKa = 4.14GVFWVASNQADD154 pKa = 3.42INTPADD160 pKa = 3.29IVDD163 pKa = 4.43RR164 pKa = 11.84DD165 pKa = 3.85PSSDD169 pKa = 3.16EE170 pKa = 4.8AIPTRR175 pKa = 11.84FPPGTVLPQGYY186 pKa = 9.38YY187 pKa = 9.56IEE189 pKa = 4.86GSGRR193 pKa = 11.84SAPNSRR199 pKa = 11.84STSRR203 pKa = 11.84APNRR207 pKa = 11.84APSAGSRR214 pKa = 11.84SRR216 pKa = 11.84ANSGNRR222 pKa = 11.84TSTPGVTPDD231 pKa = 3.56MADD234 pKa = 3.28QIASLVLAKK243 pKa = 10.41LGKK246 pKa = 10.07DD247 pKa = 2.95ATKK250 pKa = 9.49PQQVTKK256 pKa = 9.36QTAKK260 pKa = 10.33EE261 pKa = 4.03VRR263 pKa = 11.84QKK265 pKa = 10.72ILNKK269 pKa = 9.23PRR271 pKa = 11.84QKK273 pKa = 10.46RR274 pKa = 11.84SPNKK278 pKa = 9.02QCTVQQCFGKK288 pKa = 10.48RR289 pKa = 11.84GPNQNFGGGEE299 pKa = 3.99MLKK302 pKa = 10.67LGTSDD307 pKa = 3.38PQFPILAEE315 pKa = 4.05LAPTAGAFFFGSRR328 pKa = 11.84LEE330 pKa = 4.03LAKK333 pKa = 10.7VQNLSGNPDD342 pKa = 3.48EE343 pKa = 4.7PQKK346 pKa = 11.04DD347 pKa = 4.22VYY349 pKa = 9.8EE350 pKa = 3.97LRR352 pKa = 11.84YY353 pKa = 10.38NGAIRR358 pKa = 11.84FDD360 pKa = 3.73STLSGFEE367 pKa = 4.32TIMKK371 pKa = 9.76VLNQNLNAYY380 pKa = 7.22QHH382 pKa = 6.17QEE384 pKa = 3.85DD385 pKa = 3.9GMMNISPKK393 pKa = 8.82PQRR396 pKa = 11.84QRR398 pKa = 11.84GQKK401 pKa = 9.61NGQVEE406 pKa = 4.3NDD408 pKa = 3.75NVSVAAPKK416 pKa = 10.71SRR418 pKa = 11.84VQQNKK423 pKa = 9.07SRR425 pKa = 11.84EE426 pKa = 4.15LTAEE430 pKa = 4.92DD431 pKa = 3.26ISLLKK436 pKa = 11.01KK437 pKa = 9.61MDD439 pKa = 3.6EE440 pKa = 4.96PYY442 pKa = 10.5TEE444 pKa = 4.35DD445 pKa = 3.07TSEE448 pKa = 3.68II449 pKa = 3.91
Molecular weight: 49.51 kDa
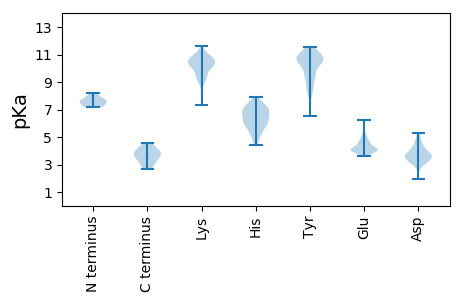
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2872 |
20 |
1349 |
359.0 |
40.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.71 ± 0.51 | 3.099 ± 0.817 |
4.735 ± 0.327 | 3.238 ± 0.512 |
5.571 ± 0.599 | 6.755 ± 0.414 |
1.323 ± 0.231 | 6.128 ± 0.748 |
4.074 ± 0.559 | 9.192 ± 1.265 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.671 ± 0.364 | 6.685 ± 0.934 |
4.701 ± 0.684 | 4.352 ± 0.817 |
3.969 ± 0.804 | 8.217 ± 0.397 |
6.929 ± 0.423 | 7.173 ± 0.583 |
1.288 ± 0.236 | 5.188 ± 0.813 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
