
Auraticoccus monumenti
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Propionibacteriaceae; Auraticoccus
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
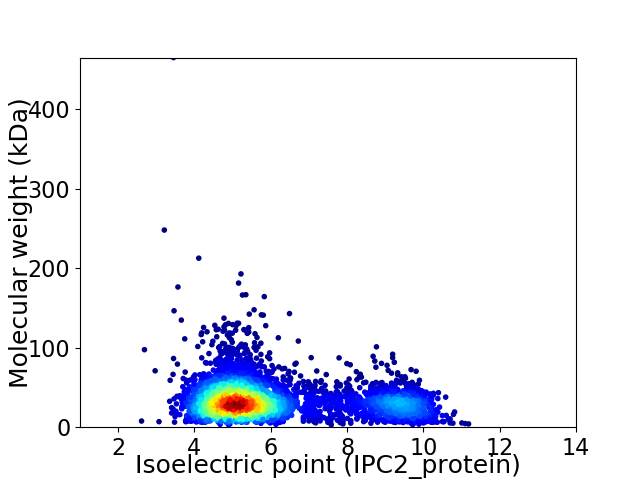
Virtual 2D-PAGE plot for 4049 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
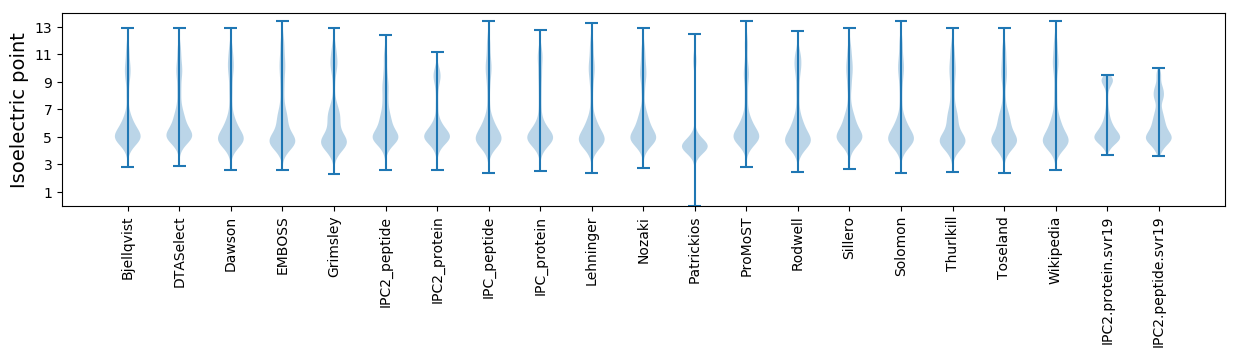
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G6V003|A0A1G6V003_9ACTN NADPH:quinone reductase OS=Auraticoccus monumenti OX=675864 GN=SAMN04489747_1007 PE=4 SV=1
MM1 pKa = 7.18VSPRR5 pKa = 11.84HH6 pKa = 5.5RR7 pKa = 11.84LHH9 pKa = 6.01RR10 pKa = 11.84TAAALVGGLLVTTTLLAPAAASPLDD35 pKa = 3.61IPHH38 pKa = 7.27DD39 pKa = 4.48FSAGPDD45 pKa = 2.92GWFSYY50 pKa = 11.31GPGATTEE57 pKa = 4.16VTDD60 pKa = 5.41GEE62 pKa = 4.93LCTSLPAHH70 pKa = 6.75SGNPWDD76 pKa = 3.89VAVAHH81 pKa = 6.1NAVGLEE87 pKa = 3.65AGTTYY92 pKa = 11.17DD93 pKa = 3.35VAFTASSTAEE103 pKa = 3.56IAIRR107 pKa = 11.84AQVGINAEE115 pKa = 4.27PYY117 pKa = 9.35PGVHH121 pKa = 6.85AEE123 pKa = 4.63DD124 pKa = 3.86LTITATPEE132 pKa = 3.5EE133 pKa = 4.61HH134 pKa = 6.83SFSFSFTSDD143 pKa = 2.51VDD145 pKa = 3.66AEE147 pKa = 4.07AAEE150 pKa = 4.44TGGQLSFQIGNQGEE164 pKa = 4.55PFTFCLDD171 pKa = 3.67DD172 pKa = 4.27FSLTEE177 pKa = 4.0AGATEE182 pKa = 4.23EE183 pKa = 4.1QLPALDD189 pKa = 3.9VPPFFTTGNLTPSTTVEE206 pKa = 4.05EE207 pKa = 4.85GVLCTTVPAGTTNTYY222 pKa = 10.77DD223 pKa = 4.37AIVGVNGIPVQEE235 pKa = 4.51GGSYY239 pKa = 9.26TLSFLASADD248 pKa = 3.61VASDD252 pKa = 2.95ITVVVGEE259 pKa = 4.18NGGDD263 pKa = 3.41YY264 pKa = 10.58RR265 pKa = 11.84QAFSQAVALDD275 pKa = 3.58SAEE278 pKa = 3.99PTRR281 pKa = 11.84YY282 pKa = 9.9SYY284 pKa = 11.56EE285 pKa = 4.02FTSDD289 pKa = 3.21LAFPASTGDD298 pKa = 3.69GAPEE302 pKa = 3.96GQVAFQLGGGAEE314 pKa = 3.95AFEE317 pKa = 5.07LCLSEE322 pKa = 4.62PSLALEE328 pKa = 4.34VAAGGNLVPNGDD340 pKa = 4.11FGSGALDD347 pKa = 3.97PFTAPEE353 pKa = 4.39GVAVDD358 pKa = 4.53LSSGAACLTVPDD370 pKa = 4.46TLGQYY375 pKa = 10.7DD376 pKa = 4.05GLVLNGLAVEE386 pKa = 4.78EE387 pKa = 4.65GQTYY391 pKa = 8.33TLTFDD396 pKa = 4.5ASADD400 pKa = 3.41PGKK403 pKa = 8.58TVRR406 pKa = 11.84ALIGEE411 pKa = 4.26NGGSYY416 pKa = 9.39GTVLDD421 pKa = 4.12EE422 pKa = 5.65DD423 pKa = 4.08VALTAEE429 pKa = 4.56TTGHH433 pKa = 6.59EE434 pKa = 4.3LTFVASAGYY443 pKa = 8.49PAATVDD449 pKa = 3.71PGDD452 pKa = 4.13GPFEE456 pKa = 4.27GQLAFQVGGRR466 pKa = 11.84GAFDD470 pKa = 3.47FCIDD474 pKa = 4.75DD475 pKa = 3.5ISLVEE480 pKa = 4.8SDD482 pKa = 4.61TPPPPYY488 pKa = 10.24EE489 pKa = 4.66PEE491 pKa = 3.73TGPRR495 pKa = 11.84IRR497 pKa = 11.84VNQVGYY503 pKa = 10.65LPDD506 pKa = 4.04GPKK509 pKa = 9.91QATLVTDD516 pKa = 3.22ATEE519 pKa = 4.01PLEE522 pKa = 4.15YY523 pKa = 10.23EE524 pKa = 4.32VLADD528 pKa = 4.57GIVVHH533 pKa = 6.7TGQTVPAGLDD543 pKa = 3.22EE544 pKa = 4.3TAGTDD549 pKa = 3.28VHH551 pKa = 6.77EE552 pKa = 5.44LDD554 pKa = 4.31FSDD557 pKa = 3.89YY558 pKa = 11.07RR559 pKa = 11.84PDD561 pKa = 3.61VVDD564 pKa = 4.02ADD566 pKa = 4.07AVHH569 pKa = 6.93EE570 pKa = 4.12IAVDD574 pKa = 3.94GEE576 pKa = 4.57VSFEE580 pKa = 3.82FAVRR584 pKa = 11.84DD585 pKa = 4.0DD586 pKa = 5.25LYY588 pKa = 11.06QQLRR592 pKa = 11.84ADD594 pKa = 3.97ALNYY598 pKa = 8.93FYY600 pKa = 10.23PARR603 pKa = 11.84SGIAIDD609 pKa = 3.79GGIMDD614 pKa = 4.99GQPDD618 pKa = 3.68AGEE621 pKa = 4.04YY622 pKa = 9.68TRR624 pKa = 11.84DD625 pKa = 3.48AGHH628 pKa = 6.63VEE630 pKa = 4.7DD631 pKa = 5.55PDD633 pKa = 4.02SSLVADD639 pKa = 3.92SPNRR643 pKa = 11.84GDD645 pKa = 3.2VDD647 pKa = 4.01VRR649 pKa = 11.84CLSPEE654 pKa = 4.0TEE656 pKa = 3.79GDD658 pKa = 3.24YY659 pKa = 10.44WKK661 pKa = 11.26YY662 pKa = 11.28GDD664 pKa = 3.85WSCDD668 pKa = 3.2YY669 pKa = 10.37TLDD672 pKa = 4.87VVGGWYY678 pKa = 10.02DD679 pKa = 3.33AGDD682 pKa = 3.49HH683 pKa = 6.07GKK685 pKa = 10.25YY686 pKa = 9.61VVNGGISVGQVLSTYY701 pKa = 10.28EE702 pKa = 3.85RR703 pKa = 11.84TLYY706 pKa = 10.95SPTGTGSALGDD717 pKa = 3.45GSLNIPEE724 pKa = 4.71SDD726 pKa = 3.44NGVPDD731 pKa = 4.58PLDD734 pKa = 3.42EE735 pKa = 5.14ARR737 pKa = 11.84WEE739 pKa = 4.21LEE741 pKa = 3.57WMLSMQAPDD750 pKa = 3.96GAQHH754 pKa = 6.92AGMVHH759 pKa = 6.53HH760 pKa = 7.35KK761 pKa = 10.18IADD764 pKa = 3.71VDD766 pKa = 3.8WTGLPLLPSEE776 pKa = 5.14DD777 pKa = 3.5PQEE780 pKa = 4.78RR781 pKa = 11.84YY782 pKa = 8.03LHH784 pKa = 6.3RR785 pKa = 11.84PSTAATLNLAAVAAQGARR803 pKa = 11.84LWAPYY808 pKa = 9.84DD809 pKa = 3.35QAFADD814 pKa = 4.24EE815 pKa = 4.73LLAAARR821 pKa = 11.84VAWDD825 pKa = 3.66AAVATPDD832 pKa = 3.78LFEE835 pKa = 4.86PAPNADD841 pKa = 4.23PSPGSGPYY849 pKa = 10.44DD850 pKa = 4.07DD851 pKa = 6.27DD852 pKa = 3.91EE853 pKa = 6.05VGDD856 pKa = 3.83EE857 pKa = 4.94FYY859 pKa = 10.43WAAAEE864 pKa = 4.36LFLTTGEE871 pKa = 4.18EE872 pKa = 4.05AFEE875 pKa = 4.02EE876 pKa = 4.55HH877 pKa = 6.81VLGSPFHH884 pKa = 6.25TADD887 pKa = 2.6IWSPSGFSWGSTAALGRR904 pKa = 11.84MDD906 pKa = 4.73LATVEE911 pKa = 4.16SDD913 pKa = 3.54LPGRR917 pKa = 11.84DD918 pKa = 3.53DD919 pKa = 3.33VRR921 pKa = 11.84ASVVEE926 pKa = 4.03GADD929 pKa = 4.6RR930 pKa = 11.84YY931 pKa = 10.9LGWQQAQPFGTAYY944 pKa = 9.71PGNEE948 pKa = 3.68GVYY951 pKa = 9.77EE952 pKa = 4.1WGSNSAVLNNQVVIATAFDD971 pKa = 3.67LTSDD975 pKa = 3.41QVYY978 pKa = 11.11ADD980 pKa = 4.09AVLEE984 pKa = 4.21AMDD987 pKa = 4.04YY988 pKa = 11.26LLGRR992 pKa = 11.84NALNNSYY999 pKa = 8.48ITGYY1003 pKa = 11.22GDD1005 pKa = 3.51VFSDD1009 pKa = 4.0DD1010 pKa = 3.23QHH1012 pKa = 7.8SRR1014 pKa = 11.84WFAHH1018 pKa = 6.99SIDD1021 pKa = 4.46PDD1023 pKa = 3.95LPNPPRR1029 pKa = 11.84GSVAGGPNSDD1039 pKa = 3.25VGTWDD1044 pKa = 3.47PVISGLYY1051 pKa = 10.2DD1052 pKa = 3.61EE1053 pKa = 5.9EE1054 pKa = 4.93RR1055 pKa = 11.84MCAPQRR1061 pKa = 11.84CYY1063 pKa = 11.08LDD1065 pKa = 5.99DD1066 pKa = 3.88IQSWSTNEE1074 pKa = 3.46ITVNWNSALSWVASFVADD1092 pKa = 3.56QQAGDD1097 pKa = 3.73EE1098 pKa = 4.39SGAGQVVDD1106 pKa = 4.69VVDD1109 pKa = 4.31HH1110 pKa = 6.97PEE1112 pKa = 3.99DD1113 pKa = 3.4MTVEE1117 pKa = 4.07EE1118 pKa = 5.22GEE1120 pKa = 4.44DD1121 pKa = 3.51AVFTASASGDD1131 pKa = 3.82PVPDD1135 pKa = 3.68VQWQRR1140 pKa = 11.84LVDD1143 pKa = 4.26GVWTDD1148 pKa = 2.93VDD1150 pKa = 3.8GAEE1153 pKa = 4.19EE1154 pKa = 3.99EE1155 pKa = 4.77TFTFEE1160 pKa = 5.72AALEE1164 pKa = 4.27DD1165 pKa = 4.38DD1166 pKa = 4.34GALVRR1171 pKa = 11.84AYY1173 pKa = 10.03FSNGFGGFFTDD1184 pKa = 4.03PATLSVTDD1192 pKa = 4.13AEE1194 pKa = 4.51PRR1196 pKa = 11.84PTPGPSDD1203 pKa = 3.92EE1204 pKa = 5.26PDD1206 pKa = 4.35DD1207 pKa = 4.22GTPDD1211 pKa = 4.41DD1212 pKa = 5.62GGAPDD1217 pKa = 5.47DD1218 pKa = 4.62GASPGAGGLPEE1229 pKa = 5.94DD1230 pKa = 4.73GGTPSEE1236 pKa = 4.46PGAGGGPTQNLSDD1249 pKa = 4.23TGPSDD1254 pKa = 3.98LVLPALLTAVLLLAVGSVVVARR1276 pKa = 11.84GRR1278 pKa = 11.84RR1279 pKa = 11.84QQDD1282 pKa = 3.12EE1283 pKa = 4.41LGG1285 pKa = 3.65
MM1 pKa = 7.18VSPRR5 pKa = 11.84HH6 pKa = 5.5RR7 pKa = 11.84LHH9 pKa = 6.01RR10 pKa = 11.84TAAALVGGLLVTTTLLAPAAASPLDD35 pKa = 3.61IPHH38 pKa = 7.27DD39 pKa = 4.48FSAGPDD45 pKa = 2.92GWFSYY50 pKa = 11.31GPGATTEE57 pKa = 4.16VTDD60 pKa = 5.41GEE62 pKa = 4.93LCTSLPAHH70 pKa = 6.75SGNPWDD76 pKa = 3.89VAVAHH81 pKa = 6.1NAVGLEE87 pKa = 3.65AGTTYY92 pKa = 11.17DD93 pKa = 3.35VAFTASSTAEE103 pKa = 3.56IAIRR107 pKa = 11.84AQVGINAEE115 pKa = 4.27PYY117 pKa = 9.35PGVHH121 pKa = 6.85AEE123 pKa = 4.63DD124 pKa = 3.86LTITATPEE132 pKa = 3.5EE133 pKa = 4.61HH134 pKa = 6.83SFSFSFTSDD143 pKa = 2.51VDD145 pKa = 3.66AEE147 pKa = 4.07AAEE150 pKa = 4.44TGGQLSFQIGNQGEE164 pKa = 4.55PFTFCLDD171 pKa = 3.67DD172 pKa = 4.27FSLTEE177 pKa = 4.0AGATEE182 pKa = 4.23EE183 pKa = 4.1QLPALDD189 pKa = 3.9VPPFFTTGNLTPSTTVEE206 pKa = 4.05EE207 pKa = 4.85GVLCTTVPAGTTNTYY222 pKa = 10.77DD223 pKa = 4.37AIVGVNGIPVQEE235 pKa = 4.51GGSYY239 pKa = 9.26TLSFLASADD248 pKa = 3.61VASDD252 pKa = 2.95ITVVVGEE259 pKa = 4.18NGGDD263 pKa = 3.41YY264 pKa = 10.58RR265 pKa = 11.84QAFSQAVALDD275 pKa = 3.58SAEE278 pKa = 3.99PTRR281 pKa = 11.84YY282 pKa = 9.9SYY284 pKa = 11.56EE285 pKa = 4.02FTSDD289 pKa = 3.21LAFPASTGDD298 pKa = 3.69GAPEE302 pKa = 3.96GQVAFQLGGGAEE314 pKa = 3.95AFEE317 pKa = 5.07LCLSEE322 pKa = 4.62PSLALEE328 pKa = 4.34VAAGGNLVPNGDD340 pKa = 4.11FGSGALDD347 pKa = 3.97PFTAPEE353 pKa = 4.39GVAVDD358 pKa = 4.53LSSGAACLTVPDD370 pKa = 4.46TLGQYY375 pKa = 10.7DD376 pKa = 4.05GLVLNGLAVEE386 pKa = 4.78EE387 pKa = 4.65GQTYY391 pKa = 8.33TLTFDD396 pKa = 4.5ASADD400 pKa = 3.41PGKK403 pKa = 8.58TVRR406 pKa = 11.84ALIGEE411 pKa = 4.26NGGSYY416 pKa = 9.39GTVLDD421 pKa = 4.12EE422 pKa = 5.65DD423 pKa = 4.08VALTAEE429 pKa = 4.56TTGHH433 pKa = 6.59EE434 pKa = 4.3LTFVASAGYY443 pKa = 8.49PAATVDD449 pKa = 3.71PGDD452 pKa = 4.13GPFEE456 pKa = 4.27GQLAFQVGGRR466 pKa = 11.84GAFDD470 pKa = 3.47FCIDD474 pKa = 4.75DD475 pKa = 3.5ISLVEE480 pKa = 4.8SDD482 pKa = 4.61TPPPPYY488 pKa = 10.24EE489 pKa = 4.66PEE491 pKa = 3.73TGPRR495 pKa = 11.84IRR497 pKa = 11.84VNQVGYY503 pKa = 10.65LPDD506 pKa = 4.04GPKK509 pKa = 9.91QATLVTDD516 pKa = 3.22ATEE519 pKa = 4.01PLEE522 pKa = 4.15YY523 pKa = 10.23EE524 pKa = 4.32VLADD528 pKa = 4.57GIVVHH533 pKa = 6.7TGQTVPAGLDD543 pKa = 3.22EE544 pKa = 4.3TAGTDD549 pKa = 3.28VHH551 pKa = 6.77EE552 pKa = 5.44LDD554 pKa = 4.31FSDD557 pKa = 3.89YY558 pKa = 11.07RR559 pKa = 11.84PDD561 pKa = 3.61VVDD564 pKa = 4.02ADD566 pKa = 4.07AVHH569 pKa = 6.93EE570 pKa = 4.12IAVDD574 pKa = 3.94GEE576 pKa = 4.57VSFEE580 pKa = 3.82FAVRR584 pKa = 11.84DD585 pKa = 4.0DD586 pKa = 5.25LYY588 pKa = 11.06QQLRR592 pKa = 11.84ADD594 pKa = 3.97ALNYY598 pKa = 8.93FYY600 pKa = 10.23PARR603 pKa = 11.84SGIAIDD609 pKa = 3.79GGIMDD614 pKa = 4.99GQPDD618 pKa = 3.68AGEE621 pKa = 4.04YY622 pKa = 9.68TRR624 pKa = 11.84DD625 pKa = 3.48AGHH628 pKa = 6.63VEE630 pKa = 4.7DD631 pKa = 5.55PDD633 pKa = 4.02SSLVADD639 pKa = 3.92SPNRR643 pKa = 11.84GDD645 pKa = 3.2VDD647 pKa = 4.01VRR649 pKa = 11.84CLSPEE654 pKa = 4.0TEE656 pKa = 3.79GDD658 pKa = 3.24YY659 pKa = 10.44WKK661 pKa = 11.26YY662 pKa = 11.28GDD664 pKa = 3.85WSCDD668 pKa = 3.2YY669 pKa = 10.37TLDD672 pKa = 4.87VVGGWYY678 pKa = 10.02DD679 pKa = 3.33AGDD682 pKa = 3.49HH683 pKa = 6.07GKK685 pKa = 10.25YY686 pKa = 9.61VVNGGISVGQVLSTYY701 pKa = 10.28EE702 pKa = 3.85RR703 pKa = 11.84TLYY706 pKa = 10.95SPTGTGSALGDD717 pKa = 3.45GSLNIPEE724 pKa = 4.71SDD726 pKa = 3.44NGVPDD731 pKa = 4.58PLDD734 pKa = 3.42EE735 pKa = 5.14ARR737 pKa = 11.84WEE739 pKa = 4.21LEE741 pKa = 3.57WMLSMQAPDD750 pKa = 3.96GAQHH754 pKa = 6.92AGMVHH759 pKa = 6.53HH760 pKa = 7.35KK761 pKa = 10.18IADD764 pKa = 3.71VDD766 pKa = 3.8WTGLPLLPSEE776 pKa = 5.14DD777 pKa = 3.5PQEE780 pKa = 4.78RR781 pKa = 11.84YY782 pKa = 8.03LHH784 pKa = 6.3RR785 pKa = 11.84PSTAATLNLAAVAAQGARR803 pKa = 11.84LWAPYY808 pKa = 9.84DD809 pKa = 3.35QAFADD814 pKa = 4.24EE815 pKa = 4.73LLAAARR821 pKa = 11.84VAWDD825 pKa = 3.66AAVATPDD832 pKa = 3.78LFEE835 pKa = 4.86PAPNADD841 pKa = 4.23PSPGSGPYY849 pKa = 10.44DD850 pKa = 4.07DD851 pKa = 6.27DD852 pKa = 3.91EE853 pKa = 6.05VGDD856 pKa = 3.83EE857 pKa = 4.94FYY859 pKa = 10.43WAAAEE864 pKa = 4.36LFLTTGEE871 pKa = 4.18EE872 pKa = 4.05AFEE875 pKa = 4.02EE876 pKa = 4.55HH877 pKa = 6.81VLGSPFHH884 pKa = 6.25TADD887 pKa = 2.6IWSPSGFSWGSTAALGRR904 pKa = 11.84MDD906 pKa = 4.73LATVEE911 pKa = 4.16SDD913 pKa = 3.54LPGRR917 pKa = 11.84DD918 pKa = 3.53DD919 pKa = 3.33VRR921 pKa = 11.84ASVVEE926 pKa = 4.03GADD929 pKa = 4.6RR930 pKa = 11.84YY931 pKa = 10.9LGWQQAQPFGTAYY944 pKa = 9.71PGNEE948 pKa = 3.68GVYY951 pKa = 9.77EE952 pKa = 4.1WGSNSAVLNNQVVIATAFDD971 pKa = 3.67LTSDD975 pKa = 3.41QVYY978 pKa = 11.11ADD980 pKa = 4.09AVLEE984 pKa = 4.21AMDD987 pKa = 4.04YY988 pKa = 11.26LLGRR992 pKa = 11.84NALNNSYY999 pKa = 8.48ITGYY1003 pKa = 11.22GDD1005 pKa = 3.51VFSDD1009 pKa = 4.0DD1010 pKa = 3.23QHH1012 pKa = 7.8SRR1014 pKa = 11.84WFAHH1018 pKa = 6.99SIDD1021 pKa = 4.46PDD1023 pKa = 3.95LPNPPRR1029 pKa = 11.84GSVAGGPNSDD1039 pKa = 3.25VGTWDD1044 pKa = 3.47PVISGLYY1051 pKa = 10.2DD1052 pKa = 3.61EE1053 pKa = 5.9EE1054 pKa = 4.93RR1055 pKa = 11.84MCAPQRR1061 pKa = 11.84CYY1063 pKa = 11.08LDD1065 pKa = 5.99DD1066 pKa = 3.88IQSWSTNEE1074 pKa = 3.46ITVNWNSALSWVASFVADD1092 pKa = 3.56QQAGDD1097 pKa = 3.73EE1098 pKa = 4.39SGAGQVVDD1106 pKa = 4.69VVDD1109 pKa = 4.31HH1110 pKa = 6.97PEE1112 pKa = 3.99DD1113 pKa = 3.4MTVEE1117 pKa = 4.07EE1118 pKa = 5.22GEE1120 pKa = 4.44DD1121 pKa = 3.51AVFTASASGDD1131 pKa = 3.82PVPDD1135 pKa = 3.68VQWQRR1140 pKa = 11.84LVDD1143 pKa = 4.26GVWTDD1148 pKa = 2.93VDD1150 pKa = 3.8GAEE1153 pKa = 4.19EE1154 pKa = 3.99EE1155 pKa = 4.77TFTFEE1160 pKa = 5.72AALEE1164 pKa = 4.27DD1165 pKa = 4.38DD1166 pKa = 4.34GALVRR1171 pKa = 11.84AYY1173 pKa = 10.03FSNGFGGFFTDD1184 pKa = 4.03PATLSVTDD1192 pKa = 4.13AEE1194 pKa = 4.51PRR1196 pKa = 11.84PTPGPSDD1203 pKa = 3.92EE1204 pKa = 5.26PDD1206 pKa = 4.35DD1207 pKa = 4.22GTPDD1211 pKa = 4.41DD1212 pKa = 5.62GGAPDD1217 pKa = 5.47DD1218 pKa = 4.62GASPGAGGLPEE1229 pKa = 5.94DD1230 pKa = 4.73GGTPSEE1236 pKa = 4.46PGAGGGPTQNLSDD1249 pKa = 4.23TGPSDD1254 pKa = 3.98LVLPALLTAVLLLAVGSVVVARR1276 pKa = 11.84GRR1278 pKa = 11.84RR1279 pKa = 11.84QQDD1282 pKa = 3.12EE1283 pKa = 4.41LGG1285 pKa = 3.65
Molecular weight: 134.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G6RNE3|A0A1G6RNE3_9ACTN Uncharacterized protein OS=Auraticoccus monumenti OX=675864 GN=SAMN04489747_0069 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.72HH17 pKa = 5.74RR18 pKa = 11.84KK19 pKa = 7.87LLKK22 pKa = 8.37RR23 pKa = 11.84TRR25 pKa = 11.84IQRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84AGKK33 pKa = 9.79
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.72HH17 pKa = 5.74RR18 pKa = 11.84KK19 pKa = 7.87LLKK22 pKa = 8.37RR23 pKa = 11.84TRR25 pKa = 11.84IQRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84AGKK33 pKa = 9.79
Molecular weight: 4.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1328520 |
29 |
4725 |
328.1 |
35.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.842 ± 0.046 | 0.681 ± 0.011 |
6.22 ± 0.034 | 5.88 ± 0.034 |
2.597 ± 0.023 | 9.397 ± 0.038 |
2.196 ± 0.021 | 2.749 ± 0.026 |
1.304 ± 0.023 | 10.766 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.7 ± 0.017 | 1.522 ± 0.02 |
6.093 ± 0.031 | 3.027 ± 0.022 |
8.059 ± 0.05 | 5.409 ± 0.029 |
6.299 ± 0.054 | 9.828 ± 0.039 |
1.63 ± 0.017 | 1.802 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
