
Labedella phragmitis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Labedella
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
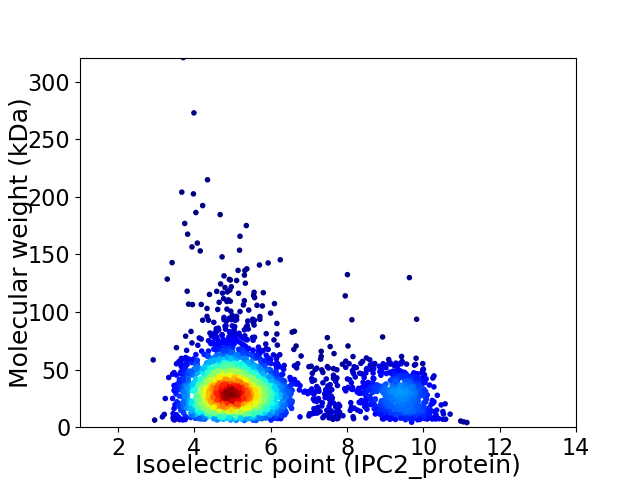
Virtual 2D-PAGE plot for 3187 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S3ZHZ2|A0A3S3ZHZ2_9MICO GNAT family N-acetyltransferase OS=Labedella phragmitis OX=2498849 GN=ELQ90_13645 PE=4 SV=1
MM1 pKa = 7.1TRR3 pKa = 11.84HH4 pKa = 5.48HH5 pKa = 7.22RR6 pKa = 11.84NRR8 pKa = 11.84LTATLAAATASLLVLAGCAGGAGSTGAAPDD38 pKa = 3.7ANAEE42 pKa = 4.29IIVGSQNEE50 pKa = 4.44PTNLDD55 pKa = 3.56QIFGGSSGVTEE66 pKa = 4.12VFTGNVYY73 pKa = 10.69EE74 pKa = 4.21GLFRR78 pKa = 11.84ITDD81 pKa = 3.73DD82 pKa = 4.8AEE84 pKa = 4.27VEE86 pKa = 4.08PLLAAEE92 pKa = 4.68TEE94 pKa = 4.21VSEE97 pKa = 6.06DD98 pKa = 2.95GLTYY102 pKa = 10.72TFTLQDD108 pKa = 3.3ATFHH112 pKa = 6.35SGAPLTAEE120 pKa = 3.75AVKK123 pKa = 10.85YY124 pKa = 10.26SLEE127 pKa = 3.97RR128 pKa = 11.84FIGEE132 pKa = 3.63EE133 pKa = 3.86SIAARR138 pKa = 11.84KK139 pKa = 8.85SQLSVIDD146 pKa = 3.63TVTAVDD152 pKa = 3.96DD153 pKa = 3.81KK154 pKa = 10.83TVEE157 pKa = 4.13VTLSSKK163 pKa = 10.83SISFTYY169 pKa = 10.02NLGYY173 pKa = 10.4VWIVNPEE180 pKa = 4.11AGDD183 pKa = 3.45LTAAADD189 pKa = 3.82GTGPYY194 pKa = 10.49SLGDD198 pKa = 3.57YY199 pKa = 10.5RR200 pKa = 11.84KK201 pKa = 9.9GDD203 pKa = 4.01SITLEE208 pKa = 4.02VNDD211 pKa = 5.51DD212 pKa = 3.62YY213 pKa = 11.6WGDD216 pKa = 3.49APANGGVVYY225 pKa = 10.11QYY227 pKa = 11.22YY228 pKa = 11.08ADD230 pKa = 3.7PTALNNALVTGAVDD244 pKa = 4.82LVTSQSNPDD253 pKa = 3.23SLAEE257 pKa = 4.12FEE259 pKa = 4.48SGDD262 pKa = 3.61YY263 pKa = 10.69EE264 pKa = 4.57IIEE267 pKa = 4.45GTSTTKK273 pKa = 10.62EE274 pKa = 3.52LLAFNDD280 pKa = 3.51RR281 pKa = 11.84VAPFDD286 pKa = 3.85DD287 pKa = 3.85VNVRR291 pKa = 11.84KK292 pKa = 9.48AVYY295 pKa = 9.83SAIDD299 pKa = 3.45RR300 pKa = 11.84EE301 pKa = 4.17QLLDD305 pKa = 4.88AIWDD309 pKa = 3.71GRR311 pKa = 11.84GQLIGSMVPPSEE323 pKa = 3.77PWYY326 pKa = 11.0LDD328 pKa = 3.57LADD331 pKa = 4.62NNPYY335 pKa = 10.66DD336 pKa = 4.35PEE338 pKa = 5.05LSADD342 pKa = 3.91LLADD346 pKa = 3.9AGYY349 pKa = 11.41ADD351 pKa = 4.58GFSFTIDD358 pKa = 3.43TPDD361 pKa = 3.39SGVHH365 pKa = 4.16STVAEE370 pKa = 4.23FLQTEE375 pKa = 4.35LAKK378 pKa = 11.04VGVTVDD384 pKa = 3.22INIITDD390 pKa = 3.59DD391 pKa = 3.08EE392 pKa = 4.66WYY394 pKa = 9.77EE395 pKa = 4.36KK396 pKa = 10.96VYY398 pKa = 10.54TDD400 pKa = 4.27HH401 pKa = 7.53DD402 pKa = 4.28FEE404 pKa = 4.72ATLQGHH410 pKa = 5.63VNDD413 pKa = 4.96RR414 pKa = 11.84DD415 pKa = 3.37INFYY419 pKa = 11.17GNPDD423 pKa = 4.29FYY425 pKa = 10.64WGYY428 pKa = 11.34DD429 pKa = 3.48DD430 pKa = 6.71AEE432 pKa = 4.43VQGWLAEE439 pKa = 4.06AEE441 pKa = 4.34QADD444 pKa = 4.06STEE447 pKa = 4.06QQTEE451 pKa = 4.68LITQVNQKK459 pKa = 9.86ISDD462 pKa = 3.87DD463 pKa = 3.84AASVWLYY470 pKa = 10.76LNPQLRR476 pKa = 11.84IVRR479 pKa = 11.84DD480 pKa = 4.5GITGVPEE487 pKa = 3.93NGLNSLFYY495 pKa = 11.03VYY497 pKa = 10.23DD498 pKa = 3.43IQEE501 pKa = 3.75AA502 pKa = 3.49
MM1 pKa = 7.1TRR3 pKa = 11.84HH4 pKa = 5.48HH5 pKa = 7.22RR6 pKa = 11.84NRR8 pKa = 11.84LTATLAAATASLLVLAGCAGGAGSTGAAPDD38 pKa = 3.7ANAEE42 pKa = 4.29IIVGSQNEE50 pKa = 4.44PTNLDD55 pKa = 3.56QIFGGSSGVTEE66 pKa = 4.12VFTGNVYY73 pKa = 10.69EE74 pKa = 4.21GLFRR78 pKa = 11.84ITDD81 pKa = 3.73DD82 pKa = 4.8AEE84 pKa = 4.27VEE86 pKa = 4.08PLLAAEE92 pKa = 4.68TEE94 pKa = 4.21VSEE97 pKa = 6.06DD98 pKa = 2.95GLTYY102 pKa = 10.72TFTLQDD108 pKa = 3.3ATFHH112 pKa = 6.35SGAPLTAEE120 pKa = 3.75AVKK123 pKa = 10.85YY124 pKa = 10.26SLEE127 pKa = 3.97RR128 pKa = 11.84FIGEE132 pKa = 3.63EE133 pKa = 3.86SIAARR138 pKa = 11.84KK139 pKa = 8.85SQLSVIDD146 pKa = 3.63TVTAVDD152 pKa = 3.96DD153 pKa = 3.81KK154 pKa = 10.83TVEE157 pKa = 4.13VTLSSKK163 pKa = 10.83SISFTYY169 pKa = 10.02NLGYY173 pKa = 10.4VWIVNPEE180 pKa = 4.11AGDD183 pKa = 3.45LTAAADD189 pKa = 3.82GTGPYY194 pKa = 10.49SLGDD198 pKa = 3.57YY199 pKa = 10.5RR200 pKa = 11.84KK201 pKa = 9.9GDD203 pKa = 4.01SITLEE208 pKa = 4.02VNDD211 pKa = 5.51DD212 pKa = 3.62YY213 pKa = 11.6WGDD216 pKa = 3.49APANGGVVYY225 pKa = 10.11QYY227 pKa = 11.22YY228 pKa = 11.08ADD230 pKa = 3.7PTALNNALVTGAVDD244 pKa = 4.82LVTSQSNPDD253 pKa = 3.23SLAEE257 pKa = 4.12FEE259 pKa = 4.48SGDD262 pKa = 3.61YY263 pKa = 10.69EE264 pKa = 4.57IIEE267 pKa = 4.45GTSTTKK273 pKa = 10.62EE274 pKa = 3.52LLAFNDD280 pKa = 3.51RR281 pKa = 11.84VAPFDD286 pKa = 3.85DD287 pKa = 3.85VNVRR291 pKa = 11.84KK292 pKa = 9.48AVYY295 pKa = 9.83SAIDD299 pKa = 3.45RR300 pKa = 11.84EE301 pKa = 4.17QLLDD305 pKa = 4.88AIWDD309 pKa = 3.71GRR311 pKa = 11.84GQLIGSMVPPSEE323 pKa = 3.77PWYY326 pKa = 11.0LDD328 pKa = 3.57LADD331 pKa = 4.62NNPYY335 pKa = 10.66DD336 pKa = 4.35PEE338 pKa = 5.05LSADD342 pKa = 3.91LLADD346 pKa = 3.9AGYY349 pKa = 11.41ADD351 pKa = 4.58GFSFTIDD358 pKa = 3.43TPDD361 pKa = 3.39SGVHH365 pKa = 4.16STVAEE370 pKa = 4.23FLQTEE375 pKa = 4.35LAKK378 pKa = 11.04VGVTVDD384 pKa = 3.22INIITDD390 pKa = 3.59DD391 pKa = 3.08EE392 pKa = 4.66WYY394 pKa = 9.77EE395 pKa = 4.36KK396 pKa = 10.96VYY398 pKa = 10.54TDD400 pKa = 4.27HH401 pKa = 7.53DD402 pKa = 4.28FEE404 pKa = 4.72ATLQGHH410 pKa = 5.63VNDD413 pKa = 4.96RR414 pKa = 11.84DD415 pKa = 3.37INFYY419 pKa = 11.17GNPDD423 pKa = 4.29FYY425 pKa = 10.64WGYY428 pKa = 11.34DD429 pKa = 3.48DD430 pKa = 6.71AEE432 pKa = 4.43VQGWLAEE439 pKa = 4.06AEE441 pKa = 4.34QADD444 pKa = 4.06STEE447 pKa = 4.06QQTEE451 pKa = 4.68LITQVNQKK459 pKa = 9.86ISDD462 pKa = 3.87DD463 pKa = 3.84AASVWLYY470 pKa = 10.76LNPQLRR476 pKa = 11.84IVRR479 pKa = 11.84DD480 pKa = 4.5GITGVPEE487 pKa = 3.93NGLNSLFYY495 pKa = 11.03VYY497 pKa = 10.23DD498 pKa = 3.43IQEE501 pKa = 3.75AA502 pKa = 3.49
Molecular weight: 54.39 kDa
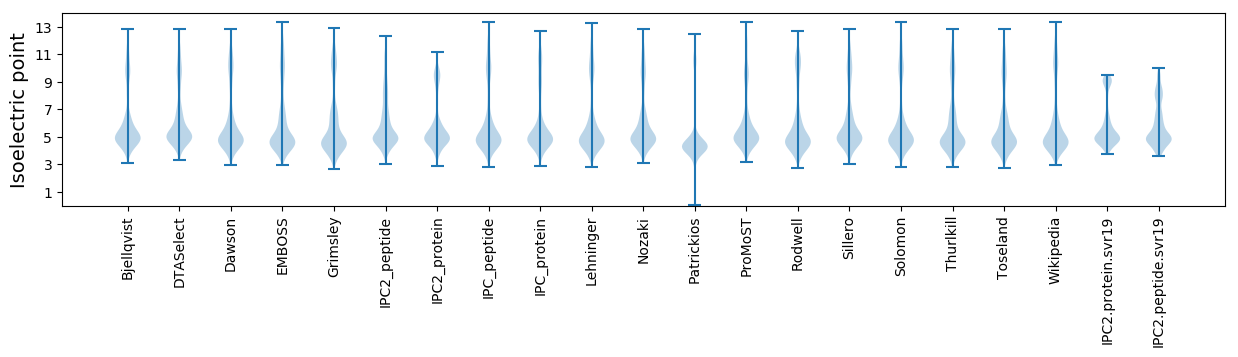
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A444PUM2|A0A444PUM2_9MICO DUF664 domain-containing protein OS=Labedella phragmitis OX=2498849 GN=ELQ90_05520 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1053170 |
32 |
3093 |
330.5 |
35.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.974 ± 0.056 | 0.459 ± 0.01 |
6.523 ± 0.045 | 5.792 ± 0.046 |
3.169 ± 0.028 | 9.033 ± 0.036 |
1.876 ± 0.021 | 4.48 ± 0.031 |
1.757 ± 0.029 | 9.816 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.645 ± 0.019 | 1.918 ± 0.025 |
5.381 ± 0.027 | 2.378 ± 0.026 |
7.334 ± 0.058 | 6.276 ± 0.034 |
6.349 ± 0.05 | 9.486 ± 0.042 |
1.411 ± 0.016 | 1.942 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
