
Diutina rugosa (Yeast) (Candida rugosa)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Saccharomycetales incertae sedis; Diutina
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
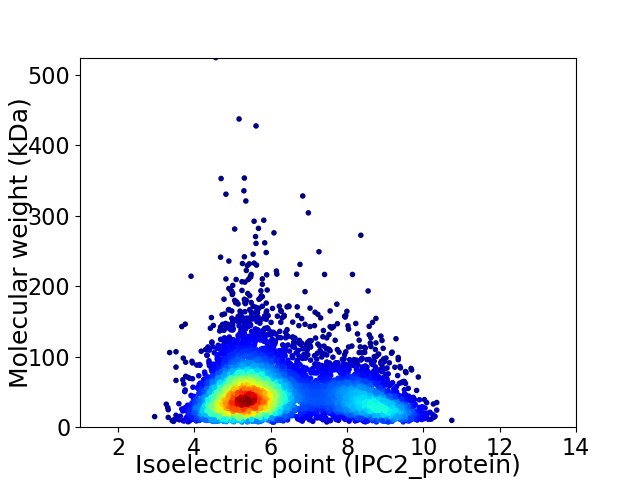
Virtual 2D-PAGE plot for 5811 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A642V1E8|A0A642V1E8_DIURU J domain-containing protein OS=Diutina rugosa OX=5481 GN=DIURU_001284 PE=3 SV=1
MM1 pKa = 7.97IIRR4 pKa = 11.84SMLLPLVAALPVLVPRR20 pKa = 11.84DD21 pKa = 3.87ADD23 pKa = 4.07DD24 pKa = 5.44DD25 pKa = 5.0DD26 pKa = 4.87NGPVTAYY33 pKa = 8.84IQPVVAVAEE42 pKa = 4.4ATTTVQPAVVYY53 pKa = 10.77VDD55 pKa = 4.49ANGNTVGGVSGAVSIDD71 pKa = 3.27DD72 pKa = 4.01SQFGTSFYY80 pKa = 7.83TTSAPPLITGDD91 pKa = 4.11EE92 pKa = 4.07NDD94 pKa = 4.17LDD96 pKa = 4.16VGGGNQDD103 pKa = 3.25NNNNNGPAAAPTAQVTNDD121 pKa = 3.81SGNGDD126 pKa = 3.84SPNTQQNNNNNVQVVEE142 pKa = 4.37TVNFGPSDD150 pKa = 3.55IVTVAATTIYY160 pKa = 10.56GPQAQDD166 pKa = 3.03NAGQQQQTSTTEE178 pKa = 3.91PQPQPTTTEE187 pKa = 3.92QQQDD191 pKa = 3.43QPTTTEE197 pKa = 3.84QPTTEE202 pKa = 4.19QPTTEE207 pKa = 4.1QPTTEE212 pKa = 4.18QQPSTEE218 pKa = 4.73PSTQPAPQPSTTQADD233 pKa = 3.66NQGDD237 pKa = 3.88NNQGNNNNNQGNNNNNSGNDD257 pKa = 3.52SNSNQGNNDD266 pKa = 3.27NNNNSDD272 pKa = 3.91NNNNYY277 pKa = 10.28NNNQGNDD284 pKa = 3.41NNQGNDD290 pKa = 3.28NNQGNDD296 pKa = 3.28NNQGNDD302 pKa = 3.23NNNNNGGSSDD312 pKa = 3.85GLLTPPVYY320 pKa = 9.8IVYY323 pKa = 10.32SPYY326 pKa = 11.32NNDD329 pKa = 3.32QSCKK333 pKa = 10.08DD334 pKa = 3.44AATVRR339 pKa = 11.84SDD341 pKa = 4.0LAIVKK346 pKa = 9.97QKK348 pKa = 10.85GINKK352 pKa = 8.22VRR354 pKa = 11.84IYY356 pKa = 11.03GSDD359 pKa = 3.05CGAFTNILPACKK371 pKa = 10.17DD372 pKa = 3.11LGLKK376 pKa = 9.56VNQGFWFQPGQGPDD390 pKa = 3.61SIDD393 pKa = 3.4GGVSEE398 pKa = 5.52FISWGQSNGWDD409 pKa = 3.1IIDD412 pKa = 3.98FVTVGNEE419 pKa = 3.6AVNDD423 pKa = 4.17GNLSPDD429 pKa = 3.39QLNGKK434 pKa = 9.13LSSVRR439 pKa = 11.84GQLQAAGYY447 pKa = 8.45SGSVITSEE455 pKa = 4.26PPISFIRR462 pKa = 11.84HH463 pKa = 6.22PEE465 pKa = 3.71LCEE468 pKa = 3.52QSDD471 pKa = 4.17FIGTNIHH478 pKa = 6.66SYY480 pKa = 10.79FNTGVAPDD488 pKa = 3.79GAGEE492 pKa = 4.01YY493 pKa = 10.53LKK495 pKa = 10.38QQNEE499 pKa = 4.37AVAQACPNKK508 pKa = 9.96KK509 pKa = 9.81IYY511 pKa = 8.77VTEE514 pKa = 3.42TGYY517 pKa = 9.57PHH519 pKa = 7.46KK520 pKa = 10.83GATNGLNVPSKK531 pKa = 9.8EE532 pKa = 3.82NQKK535 pKa = 10.0IVFEE539 pKa = 5.07SIWNTIGNDD548 pKa = 3.46VTVLAMWDD556 pKa = 4.46DD557 pKa = 3.84YY558 pKa = 10.85WKK560 pKa = 11.19NPGPYY565 pKa = 10.11GIEE568 pKa = 4.0QYY570 pKa = 10.78FGALDD575 pKa = 3.85LFSS578 pKa = 4.27
MM1 pKa = 7.97IIRR4 pKa = 11.84SMLLPLVAALPVLVPRR20 pKa = 11.84DD21 pKa = 3.87ADD23 pKa = 4.07DD24 pKa = 5.44DD25 pKa = 5.0DD26 pKa = 4.87NGPVTAYY33 pKa = 8.84IQPVVAVAEE42 pKa = 4.4ATTTVQPAVVYY53 pKa = 10.77VDD55 pKa = 4.49ANGNTVGGVSGAVSIDD71 pKa = 3.27DD72 pKa = 4.01SQFGTSFYY80 pKa = 7.83TTSAPPLITGDD91 pKa = 4.11EE92 pKa = 4.07NDD94 pKa = 4.17LDD96 pKa = 4.16VGGGNQDD103 pKa = 3.25NNNNNGPAAAPTAQVTNDD121 pKa = 3.81SGNGDD126 pKa = 3.84SPNTQQNNNNNVQVVEE142 pKa = 4.37TVNFGPSDD150 pKa = 3.55IVTVAATTIYY160 pKa = 10.56GPQAQDD166 pKa = 3.03NAGQQQQTSTTEE178 pKa = 3.91PQPQPTTTEE187 pKa = 3.92QQQDD191 pKa = 3.43QPTTTEE197 pKa = 3.84QPTTEE202 pKa = 4.19QPTTEE207 pKa = 4.1QPTTEE212 pKa = 4.18QQPSTEE218 pKa = 4.73PSTQPAPQPSTTQADD233 pKa = 3.66NQGDD237 pKa = 3.88NNQGNNNNNQGNNNNNSGNDD257 pKa = 3.52SNSNQGNNDD266 pKa = 3.27NNNNSDD272 pKa = 3.91NNNNYY277 pKa = 10.28NNNQGNDD284 pKa = 3.41NNQGNDD290 pKa = 3.28NNQGNDD296 pKa = 3.28NNQGNDD302 pKa = 3.23NNNNNGGSSDD312 pKa = 3.85GLLTPPVYY320 pKa = 9.8IVYY323 pKa = 10.32SPYY326 pKa = 11.32NNDD329 pKa = 3.32QSCKK333 pKa = 10.08DD334 pKa = 3.44AATVRR339 pKa = 11.84SDD341 pKa = 4.0LAIVKK346 pKa = 9.97QKK348 pKa = 10.85GINKK352 pKa = 8.22VRR354 pKa = 11.84IYY356 pKa = 11.03GSDD359 pKa = 3.05CGAFTNILPACKK371 pKa = 10.17DD372 pKa = 3.11LGLKK376 pKa = 9.56VNQGFWFQPGQGPDD390 pKa = 3.61SIDD393 pKa = 3.4GGVSEE398 pKa = 5.52FISWGQSNGWDD409 pKa = 3.1IIDD412 pKa = 3.98FVTVGNEE419 pKa = 3.6AVNDD423 pKa = 4.17GNLSPDD429 pKa = 3.39QLNGKK434 pKa = 9.13LSSVRR439 pKa = 11.84GQLQAAGYY447 pKa = 8.45SGSVITSEE455 pKa = 4.26PPISFIRR462 pKa = 11.84HH463 pKa = 6.22PEE465 pKa = 3.71LCEE468 pKa = 3.52QSDD471 pKa = 4.17FIGTNIHH478 pKa = 6.66SYY480 pKa = 10.79FNTGVAPDD488 pKa = 3.79GAGEE492 pKa = 4.01YY493 pKa = 10.53LKK495 pKa = 10.38QQNEE499 pKa = 4.37AVAQACPNKK508 pKa = 9.96KK509 pKa = 9.81IYY511 pKa = 8.77VTEE514 pKa = 3.42TGYY517 pKa = 9.57PHH519 pKa = 7.46KK520 pKa = 10.83GATNGLNVPSKK531 pKa = 9.8EE532 pKa = 3.82NQKK535 pKa = 10.0IVFEE539 pKa = 5.07SIWNTIGNDD548 pKa = 3.46VTVLAMWDD556 pKa = 4.46DD557 pKa = 3.84YY558 pKa = 10.85WKK560 pKa = 11.19NPGPYY565 pKa = 10.11GIEE568 pKa = 4.0QYY570 pKa = 10.78FGALDD575 pKa = 3.85LFSS578 pKa = 4.27
Molecular weight: 61.48 kDa
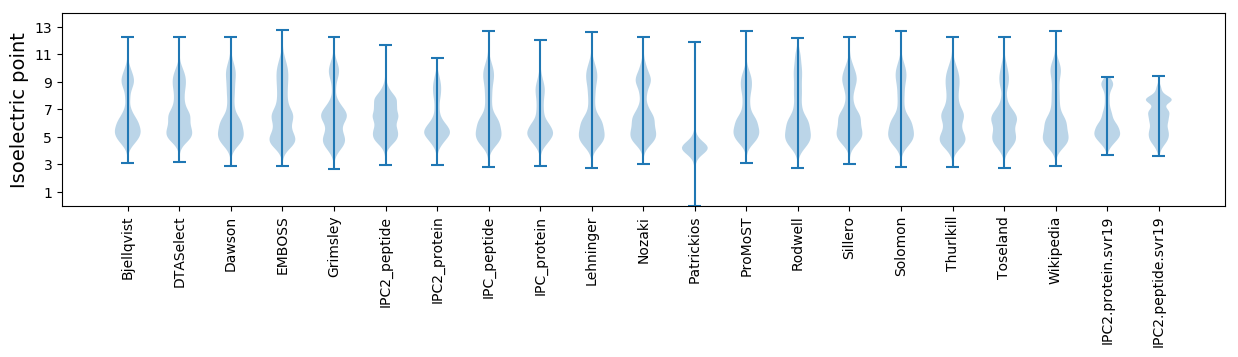
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A642URN3|A0A642URN3_DIURU Uncharacterized protein OS=Diutina rugosa OX=5481 GN=DIURU_001983 PE=3 SV=1
MM1 pKa = 7.13VFSKK5 pKa = 10.12FKK7 pKa = 11.17ANIVDD12 pKa = 3.56EE13 pKa = 4.79HH14 pKa = 6.81NGSGLISKK22 pKa = 7.86RR23 pKa = 11.84TRR25 pKa = 11.84VVSARR30 pKa = 11.84GGASRR35 pKa = 11.84TASPEE40 pKa = 3.49APRR43 pKa = 11.84YY44 pKa = 9.04LDD46 pKa = 3.34HH47 pKa = 7.28HH48 pKa = 6.92AAPPAAVRR56 pKa = 11.84TQSQQLYY63 pKa = 8.28SQPHH67 pKa = 5.49HH68 pKa = 6.3HH69 pKa = 7.15HH70 pKa = 7.29PPGSWIDD77 pKa = 3.6PRR79 pKa = 11.84ASAQRR84 pKa = 11.84STPPRR89 pKa = 11.84ASAQRR94 pKa = 11.84STPPSSSAASSVFSNPAPARR114 pKa = 11.84RR115 pKa = 11.84PSVTYY120 pKa = 9.76RR121 pKa = 11.84HH122 pKa = 6.16SAPSVKK128 pKa = 10.02VASSEE133 pKa = 4.11SSSPVSEE140 pKa = 4.84HH141 pKa = 5.38SAEE144 pKa = 4.71AIFDD148 pKa = 3.72SAPPRR153 pKa = 11.84KK154 pKa = 9.72SLLKK158 pKa = 10.37RR159 pKa = 11.84LNSQKK164 pKa = 9.29QRR166 pKa = 11.84KK167 pKa = 7.65PKK169 pKa = 10.34APRR172 pKa = 11.84TAMEE176 pKa = 4.23WAGLPPRR183 pKa = 11.84EE184 pKa = 4.12TLPPRR189 pKa = 11.84VVSSMGDD196 pKa = 3.44DD197 pKa = 5.3DD198 pKa = 6.88DD199 pKa = 6.34DD200 pKa = 6.45LPLCGPWTGRR210 pKa = 3.45
MM1 pKa = 7.13VFSKK5 pKa = 10.12FKK7 pKa = 11.17ANIVDD12 pKa = 3.56EE13 pKa = 4.79HH14 pKa = 6.81NGSGLISKK22 pKa = 7.86RR23 pKa = 11.84TRR25 pKa = 11.84VVSARR30 pKa = 11.84GGASRR35 pKa = 11.84TASPEE40 pKa = 3.49APRR43 pKa = 11.84YY44 pKa = 9.04LDD46 pKa = 3.34HH47 pKa = 7.28HH48 pKa = 6.92AAPPAAVRR56 pKa = 11.84TQSQQLYY63 pKa = 8.28SQPHH67 pKa = 5.49HH68 pKa = 6.3HH69 pKa = 7.15HH70 pKa = 7.29PPGSWIDD77 pKa = 3.6PRR79 pKa = 11.84ASAQRR84 pKa = 11.84STPPRR89 pKa = 11.84ASAQRR94 pKa = 11.84STPPSSSAASSVFSNPAPARR114 pKa = 11.84RR115 pKa = 11.84PSVTYY120 pKa = 9.76RR121 pKa = 11.84HH122 pKa = 6.16SAPSVKK128 pKa = 10.02VASSEE133 pKa = 4.11SSSPVSEE140 pKa = 4.84HH141 pKa = 5.38SAEE144 pKa = 4.71AIFDD148 pKa = 3.72SAPPRR153 pKa = 11.84KK154 pKa = 9.72SLLKK158 pKa = 10.37RR159 pKa = 11.84LNSQKK164 pKa = 9.29QRR166 pKa = 11.84KK167 pKa = 7.65PKK169 pKa = 10.34APRR172 pKa = 11.84TAMEE176 pKa = 4.23WAGLPPRR183 pKa = 11.84EE184 pKa = 4.12TLPPRR189 pKa = 11.84VVSSMGDD196 pKa = 3.44DD197 pKa = 5.3DD198 pKa = 6.88DD199 pKa = 6.34DD200 pKa = 6.45LPLCGPWTGRR210 pKa = 3.45
Molecular weight: 22.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2762008 |
67 |
4754 |
475.3 |
53.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.711 ± 0.034 | 1.116 ± 0.012 |
6.306 ± 0.025 | 6.094 ± 0.031 |
3.963 ± 0.022 | 5.685 ± 0.034 |
2.44 ± 0.013 | 5.193 ± 0.023 |
5.874 ± 0.031 | 8.817 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.137 ± 0.012 | 4.258 ± 0.02 |
5.468 ± 0.034 | 4.416 ± 0.028 |
4.961 ± 0.026 | 8.246 ± 0.039 |
5.758 ± 0.029 | 7.009 ± 0.03 |
1.22 ± 0.011 | 3.31 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
