
Spiroplasma phage SVTS2
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Plectroviridae; Suturavirus; Spiroplasma virus SVTS2
Average proteome isoelectric point is 7.55
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 13 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
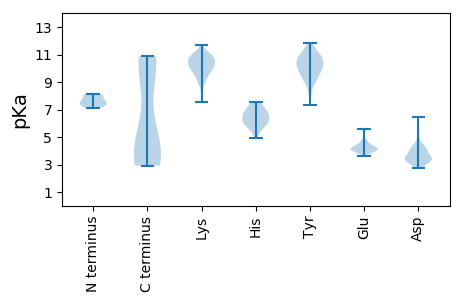
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|Q9QTH6|Q9QTH6_9VIRU ORF D OS=Spiroplasma phage SVTS2 OX=93224 PE=4 SV=1
MM1 pKa = 7.66NGFLVQLIDD10 pKa = 3.2IATGKK15 pKa = 9.95IIKK18 pKa = 9.43DD19 pKa = 3.48DD20 pKa = 4.02KK21 pKa = 11.25GNEE24 pKa = 4.43SKK26 pKa = 10.18WDD28 pKa = 3.64SYY30 pKa = 11.58TFTPVIKK37 pKa = 10.74LEE39 pKa = 4.13NGTVKK44 pKa = 9.52GTKK47 pKa = 9.82DD48 pKa = 3.18LSKK51 pKa = 11.15SKK53 pKa = 9.53WFKK56 pKa = 10.08ITDD59 pKa = 3.47EE60 pKa = 4.73NYY62 pKa = 10.99LEE64 pKa = 4.38LKK66 pKa = 10.28PYY68 pKa = 10.57LIDD71 pKa = 4.3GNLFYY76 pKa = 11.35VSLKK80 pKa = 9.6WDD82 pKa = 3.37GKK84 pKa = 9.76INIVEE89 pKa = 4.76PYY91 pKa = 8.58TEE93 pKa = 4.42NYY95 pKa = 9.88NEE97 pKa = 3.73QEE99 pKa = 4.5FINKK103 pKa = 9.33YY104 pKa = 10.24SNNSSITEE112 pKa = 4.25SNSS115 pKa = 2.92
MM1 pKa = 7.66NGFLVQLIDD10 pKa = 3.2IATGKK15 pKa = 9.95IIKK18 pKa = 9.43DD19 pKa = 3.48DD20 pKa = 4.02KK21 pKa = 11.25GNEE24 pKa = 4.43SKK26 pKa = 10.18WDD28 pKa = 3.64SYY30 pKa = 11.58TFTPVIKK37 pKa = 10.74LEE39 pKa = 4.13NGTVKK44 pKa = 9.52GTKK47 pKa = 9.82DD48 pKa = 3.18LSKK51 pKa = 11.15SKK53 pKa = 9.53WFKK56 pKa = 10.08ITDD59 pKa = 3.47EE60 pKa = 4.73NYY62 pKa = 10.99LEE64 pKa = 4.38LKK66 pKa = 10.28PYY68 pKa = 10.57LIDD71 pKa = 4.3GNLFYY76 pKa = 11.35VSLKK80 pKa = 9.6WDD82 pKa = 3.37GKK84 pKa = 9.76INIVEE89 pKa = 4.76PYY91 pKa = 8.58TEE93 pKa = 4.42NYY95 pKa = 9.88NEE97 pKa = 3.73QEE99 pKa = 4.5FINKK103 pKa = 9.33YY104 pKa = 10.24SNNSSITEE112 pKa = 4.25SNSS115 pKa = 2.92
Molecular weight: 13.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9QTH9|Q9QTH9_9VIRU ORF 1 OS=Spiroplasma phage SVTS2 OX=93224 PE=4 SV=1
MM1 pKa = 7.41NNVFIQDD8 pKa = 3.2NLTNKK13 pKa = 8.97RR14 pKa = 11.84HH15 pKa = 4.95NVFMNALNYY24 pKa = 9.36VKK26 pKa = 10.46KK27 pKa = 10.3EE28 pKa = 3.91YY29 pKa = 10.14YY30 pKa = 9.33LKK32 pKa = 10.68KK33 pKa = 10.3VFYY36 pKa = 10.81GNYY39 pKa = 8.54VKK41 pKa = 10.92NIVLPLEE48 pKa = 4.24LVNNNPRR55 pKa = 11.84NKK57 pKa = 9.72MGVKK61 pKa = 8.77NTGKK65 pKa = 9.97NDD67 pKa = 3.18KK68 pKa = 10.23KK69 pKa = 10.63LWNSRR74 pKa = 11.84IRR76 pKa = 11.84SQGNCIRR83 pKa = 11.84KK84 pKa = 9.65AYY86 pKa = 9.92EE87 pKa = 3.92NFWNCKK93 pKa = 8.53NLSFLTLTYY102 pKa = 10.78AVNEE106 pKa = 4.15KK107 pKa = 10.35DD108 pKa = 4.1VKK110 pKa = 10.52KK111 pKa = 10.81CKK113 pKa = 10.11NDD115 pKa = 3.15LKK117 pKa = 11.35LFFNNINRR125 pKa = 11.84WWNNPIRR132 pKa = 11.84SKK134 pKa = 9.03NHH136 pKa = 5.59KK137 pKa = 10.39GILKK141 pKa = 10.07YY142 pKa = 10.0MYY144 pKa = 9.1TYY146 pKa = 10.19EE147 pKa = 4.07YY148 pKa = 9.64QKK150 pKa = 10.27RR151 pKa = 11.84GAVHH155 pKa = 5.8FHH157 pKa = 6.99IILNQKK163 pKa = 9.0IPNSVVQQYY172 pKa = 7.72WKK174 pKa = 10.74HH175 pKa = 6.02GINKK179 pKa = 9.01NIKK182 pKa = 8.71VRR184 pKa = 11.84AGSNEE189 pKa = 3.88DD190 pKa = 3.38VVKK193 pKa = 10.98YY194 pKa = 8.98LAKK197 pKa = 10.85YY198 pKa = 9.05IVKK201 pKa = 8.76TANNDD206 pKa = 2.9KK207 pKa = 10.81SQNHH211 pKa = 5.95YY212 pKa = 10.84DD213 pKa = 4.01LNIKK217 pKa = 10.06AYY219 pKa = 10.18QFSKK223 pKa = 10.1NCKK226 pKa = 7.56NPKK229 pKa = 9.41VKK231 pKa = 10.71VGVIEE236 pKa = 5.62LSEE239 pKa = 3.83QDD241 pKa = 4.22LIYY244 pKa = 10.71SVKK247 pKa = 10.97DD248 pKa = 3.17NLNYY252 pKa = 10.22FSFCDD257 pKa = 3.4KK258 pKa = 10.8NGYY261 pKa = 9.71KK262 pKa = 9.74IGFSCDD268 pKa = 2.72SYY270 pKa = 11.66YY271 pKa = 11.44GLDD274 pKa = 3.31KK275 pKa = 10.79FRR277 pKa = 11.84EE278 pKa = 4.16FKK280 pKa = 10.71KK281 pKa = 10.86YY282 pKa = 10.26VSADD286 pKa = 3.24RR287 pKa = 11.84KK288 pKa = 9.98IFRR291 pKa = 11.84NLVKK295 pKa = 9.97NTNFTVNKK303 pKa = 7.85NTNLRR308 pKa = 11.84CC309 pKa = 3.83
MM1 pKa = 7.41NNVFIQDD8 pKa = 3.2NLTNKK13 pKa = 8.97RR14 pKa = 11.84HH15 pKa = 4.95NVFMNALNYY24 pKa = 9.36VKK26 pKa = 10.46KK27 pKa = 10.3EE28 pKa = 3.91YY29 pKa = 10.14YY30 pKa = 9.33LKK32 pKa = 10.68KK33 pKa = 10.3VFYY36 pKa = 10.81GNYY39 pKa = 8.54VKK41 pKa = 10.92NIVLPLEE48 pKa = 4.24LVNNNPRR55 pKa = 11.84NKK57 pKa = 9.72MGVKK61 pKa = 8.77NTGKK65 pKa = 9.97NDD67 pKa = 3.18KK68 pKa = 10.23KK69 pKa = 10.63LWNSRR74 pKa = 11.84IRR76 pKa = 11.84SQGNCIRR83 pKa = 11.84KK84 pKa = 9.65AYY86 pKa = 9.92EE87 pKa = 3.92NFWNCKK93 pKa = 8.53NLSFLTLTYY102 pKa = 10.78AVNEE106 pKa = 4.15KK107 pKa = 10.35DD108 pKa = 4.1VKK110 pKa = 10.52KK111 pKa = 10.81CKK113 pKa = 10.11NDD115 pKa = 3.15LKK117 pKa = 11.35LFFNNINRR125 pKa = 11.84WWNNPIRR132 pKa = 11.84SKK134 pKa = 9.03NHH136 pKa = 5.59KK137 pKa = 10.39GILKK141 pKa = 10.07YY142 pKa = 10.0MYY144 pKa = 9.1TYY146 pKa = 10.19EE147 pKa = 4.07YY148 pKa = 9.64QKK150 pKa = 10.27RR151 pKa = 11.84GAVHH155 pKa = 5.8FHH157 pKa = 6.99IILNQKK163 pKa = 9.0IPNSVVQQYY172 pKa = 7.72WKK174 pKa = 10.74HH175 pKa = 6.02GINKK179 pKa = 9.01NIKK182 pKa = 8.71VRR184 pKa = 11.84AGSNEE189 pKa = 3.88DD190 pKa = 3.38VVKK193 pKa = 10.98YY194 pKa = 8.98LAKK197 pKa = 10.85YY198 pKa = 9.05IVKK201 pKa = 8.76TANNDD206 pKa = 2.9KK207 pKa = 10.81SQNHH211 pKa = 5.95YY212 pKa = 10.84DD213 pKa = 4.01LNIKK217 pKa = 10.06AYY219 pKa = 10.18QFSKK223 pKa = 10.1NCKK226 pKa = 7.56NPKK229 pKa = 9.41VKK231 pKa = 10.71VGVIEE236 pKa = 5.62LSEE239 pKa = 3.83QDD241 pKa = 4.22LIYY244 pKa = 10.71SVKK247 pKa = 10.97DD248 pKa = 3.17NLNYY252 pKa = 10.22FSFCDD257 pKa = 3.4KK258 pKa = 10.8NGYY261 pKa = 9.71KK262 pKa = 9.74IGFSCDD268 pKa = 2.72SYY270 pKa = 11.66YY271 pKa = 11.44GLDD274 pKa = 3.31KK275 pKa = 10.79FRR277 pKa = 11.84EE278 pKa = 4.16FKK280 pKa = 10.71KK281 pKa = 10.86YY282 pKa = 10.26VSADD286 pKa = 3.24RR287 pKa = 11.84KK288 pKa = 9.98IFRR291 pKa = 11.84NLVKK295 pKa = 9.97NTNFTVNKK303 pKa = 7.85NTNLRR308 pKa = 11.84CC309 pKa = 3.83
Molecular weight: 36.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1810 |
52 |
394 |
139.2 |
16.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.762 ± 0.649 | 1.271 ± 0.481 |
5.304 ± 0.614 | 3.204 ± 0.521 |
8.122 ± 1.074 | 4.199 ± 0.459 |
1.271 ± 0.378 | 11.16 ± 1.184 |
9.724 ± 1.151 | 9.669 ± 0.665 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.818 ± 0.363 | 9.503 ± 1.209 |
1.989 ± 0.252 | 2.818 ± 0.413 |
2.762 ± 0.428 | 5.912 ± 0.709 |
3.923 ± 0.5 | 6.243 ± 0.764 |
1.823 ± 0.232 | 5.525 ± 0.68 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
