
Sanguibacter antarcticus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Sanguibacteraceae; Sanguibacter
Average proteome isoelectric point is 5.86
Get precalculated fractions of proteins
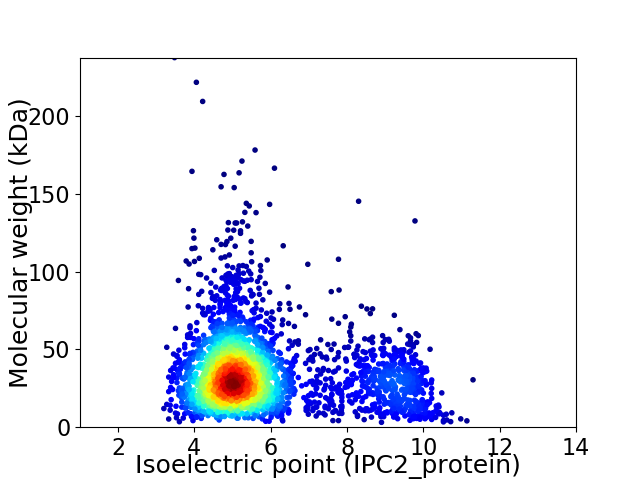
Virtual 2D-PAGE plot for 3108 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A9E8J7|A0A2A9E8J7_9MICO Two-component system sensor histidine kinase SenX3 OS=Sanguibacter antarcticus OX=372484 GN=ATL42_3210 PE=4 SV=1
MM1 pKa = 7.79EE2 pKa = 4.89SMKK5 pKa = 10.67HH6 pKa = 3.9STTPRR11 pKa = 11.84RR12 pKa = 11.84VLHH15 pKa = 6.06ALCLGSLVLALTACEE30 pKa = 4.11DD31 pKa = 3.38VRR33 pKa = 11.84AIPWPAAGTGTATDD47 pKa = 3.81SATADD52 pKa = 3.56ATPEE56 pKa = 3.96PSPVQSADD64 pKa = 3.36LEE66 pKa = 4.39ATTDD70 pKa = 3.29EE71 pKa = 4.67GTAEE75 pKa = 4.14PVEE78 pKa = 4.41VAVPDD83 pKa = 4.12DD84 pKa = 4.03VVPAPVPAALPEE96 pKa = 4.22DD97 pKa = 4.11VVPDD101 pKa = 3.91EE102 pKa = 4.87AAPAEE107 pKa = 4.24PVVVEE112 pKa = 4.64PVATAEE118 pKa = 4.43VVVSAAYY125 pKa = 9.65PDD127 pKa = 4.42DD128 pKa = 4.47AVGDD132 pKa = 3.98YY133 pKa = 10.79QLGGSYY139 pKa = 9.48TPPAGVNLVVRR150 pKa = 11.84DD151 pKa = 4.13STSKK155 pKa = 10.21PAPGLYY161 pKa = 9.99NICYY165 pKa = 9.9INGFQTQSDD174 pKa = 4.65DD175 pKa = 3.32SDD177 pKa = 3.46AWLKK181 pKa = 10.45DD182 pKa = 3.2HH183 pKa = 7.52PEE185 pKa = 4.05LVLHH189 pKa = 6.71VDD191 pKa = 4.33GEE193 pKa = 4.61PVADD197 pKa = 4.46PGWPDD202 pKa = 3.32EE203 pKa = 4.09YY204 pKa = 11.5LLDD207 pKa = 3.49VSTAAKK213 pKa = 8.24RR214 pKa = 11.84TAIADD219 pKa = 3.36IMAVTMTSCADD230 pKa = 3.19KK231 pKa = 11.2GFDD234 pKa = 5.01AIEE237 pKa = 4.05FDD239 pKa = 4.58NLDD242 pKa = 3.44SYY244 pKa = 11.5LRR246 pKa = 11.84SDD248 pKa = 4.29DD249 pKa = 5.19ALDD252 pKa = 3.82IDD254 pKa = 5.15DD255 pKa = 5.02AVATARR261 pKa = 11.84LYY263 pKa = 9.57TEE265 pKa = 4.81RR266 pKa = 11.84GHH268 pKa = 7.53ALGLEE273 pKa = 4.33VGQKK277 pKa = 8.16NTAEE281 pKa = 4.16LGSRR285 pKa = 11.84GRR287 pKa = 11.84DD288 pKa = 3.05EE289 pKa = 4.61VGFDD293 pKa = 3.04FAIAEE298 pKa = 4.12EE299 pKa = 4.33CGVYY303 pKa = 9.88EE304 pKa = 4.28EE305 pKa = 5.57CSDD308 pKa = 3.72FTDD311 pKa = 3.66VYY313 pKa = 10.14GSHH316 pKa = 6.18VFAIEE321 pKa = 3.89YY322 pKa = 10.41PEE324 pKa = 6.13DD325 pKa = 3.68GDD327 pKa = 4.16FAEE330 pKa = 5.29SCASPGRR337 pKa = 11.84PDD339 pKa = 4.29NMILRR344 pKa = 11.84DD345 pKa = 4.09LDD347 pKa = 4.07LVTPSDD353 pKa = 3.11SDD355 pKa = 3.76YY356 pKa = 11.52VYY358 pKa = 10.73EE359 pKa = 4.18RR360 pKa = 11.84CC361 pKa = 4.98
MM1 pKa = 7.79EE2 pKa = 4.89SMKK5 pKa = 10.67HH6 pKa = 3.9STTPRR11 pKa = 11.84RR12 pKa = 11.84VLHH15 pKa = 6.06ALCLGSLVLALTACEE30 pKa = 4.11DD31 pKa = 3.38VRR33 pKa = 11.84AIPWPAAGTGTATDD47 pKa = 3.81SATADD52 pKa = 3.56ATPEE56 pKa = 3.96PSPVQSADD64 pKa = 3.36LEE66 pKa = 4.39ATTDD70 pKa = 3.29EE71 pKa = 4.67GTAEE75 pKa = 4.14PVEE78 pKa = 4.41VAVPDD83 pKa = 4.12DD84 pKa = 4.03VVPAPVPAALPEE96 pKa = 4.22DD97 pKa = 4.11VVPDD101 pKa = 3.91EE102 pKa = 4.87AAPAEE107 pKa = 4.24PVVVEE112 pKa = 4.64PVATAEE118 pKa = 4.43VVVSAAYY125 pKa = 9.65PDD127 pKa = 4.42DD128 pKa = 4.47AVGDD132 pKa = 3.98YY133 pKa = 10.79QLGGSYY139 pKa = 9.48TPPAGVNLVVRR150 pKa = 11.84DD151 pKa = 4.13STSKK155 pKa = 10.21PAPGLYY161 pKa = 9.99NICYY165 pKa = 9.9INGFQTQSDD174 pKa = 4.65DD175 pKa = 3.32SDD177 pKa = 3.46AWLKK181 pKa = 10.45DD182 pKa = 3.2HH183 pKa = 7.52PEE185 pKa = 4.05LVLHH189 pKa = 6.71VDD191 pKa = 4.33GEE193 pKa = 4.61PVADD197 pKa = 4.46PGWPDD202 pKa = 3.32EE203 pKa = 4.09YY204 pKa = 11.5LLDD207 pKa = 3.49VSTAAKK213 pKa = 8.24RR214 pKa = 11.84TAIADD219 pKa = 3.36IMAVTMTSCADD230 pKa = 3.19KK231 pKa = 11.2GFDD234 pKa = 5.01AIEE237 pKa = 4.05FDD239 pKa = 4.58NLDD242 pKa = 3.44SYY244 pKa = 11.5LRR246 pKa = 11.84SDD248 pKa = 4.29DD249 pKa = 5.19ALDD252 pKa = 3.82IDD254 pKa = 5.15DD255 pKa = 5.02AVATARR261 pKa = 11.84LYY263 pKa = 9.57TEE265 pKa = 4.81RR266 pKa = 11.84GHH268 pKa = 7.53ALGLEE273 pKa = 4.33VGQKK277 pKa = 8.16NTAEE281 pKa = 4.16LGSRR285 pKa = 11.84GRR287 pKa = 11.84DD288 pKa = 3.05EE289 pKa = 4.61VGFDD293 pKa = 3.04FAIAEE298 pKa = 4.12EE299 pKa = 4.33CGVYY303 pKa = 9.88EE304 pKa = 4.28EE305 pKa = 5.57CSDD308 pKa = 3.72FTDD311 pKa = 3.66VYY313 pKa = 10.14GSHH316 pKa = 6.18VFAIEE321 pKa = 3.89YY322 pKa = 10.41PEE324 pKa = 6.13DD325 pKa = 3.68GDD327 pKa = 4.16FAEE330 pKa = 5.29SCASPGRR337 pKa = 11.84PDD339 pKa = 4.29NMILRR344 pKa = 11.84DD345 pKa = 4.09LDD347 pKa = 4.07LVTPSDD353 pKa = 3.11SDD355 pKa = 3.76YY356 pKa = 11.52VYY358 pKa = 10.73EE359 pKa = 4.18RR360 pKa = 11.84CC361 pKa = 4.98
Molecular weight: 38.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A9E3D3|A0A2A9E3D3_9MICO Ribosomal silencing factor RsfS OS=Sanguibacter antarcticus OX=372484 GN=rsfS PE=3 SV=1
MM1 pKa = 7.96PSPLLRR7 pKa = 11.84LARR10 pKa = 11.84SASRR14 pKa = 11.84QHH16 pKa = 5.82STSSPSSPRR25 pKa = 11.84LRR27 pKa = 11.84PSPPSPPSVAVRR39 pKa = 11.84RR40 pKa = 11.84PPDD43 pKa = 4.07GPPPAPQFSTPLCPTRR59 pKa = 11.84SSPPPQRR66 pKa = 11.84RR67 pKa = 11.84PALPRR72 pKa = 11.84PTRR75 pKa = 11.84SGPRR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84AAAGHH86 pKa = 5.53EE87 pKa = 4.02RR88 pKa = 11.84RR89 pKa = 11.84ARR91 pKa = 11.84RR92 pKa = 11.84PGTPRR97 pKa = 11.84RR98 pKa = 11.84LTTTRR103 pKa = 11.84LPRR106 pKa = 11.84STWSPLSRR114 pKa = 11.84RR115 pKa = 11.84SHH117 pKa = 6.5HH118 pKa = 6.28GRR120 pKa = 11.84LPTTPLHH127 pKa = 6.63SPLAHH132 pKa = 6.98AGHH135 pKa = 7.32AVQAARR141 pKa = 11.84RR142 pKa = 11.84ARR144 pKa = 11.84PPSRR148 pKa = 11.84RR149 pKa = 11.84LHH151 pKa = 6.42RR152 pKa = 11.84AFQRR156 pKa = 11.84HH157 pKa = 5.07RR158 pKa = 11.84ALRR161 pKa = 11.84WLLLSRR167 pKa = 11.84LSSSSRR173 pKa = 11.84LSRR176 pKa = 11.84RR177 pKa = 11.84RR178 pKa = 11.84PLSRR182 pKa = 11.84RR183 pKa = 11.84SPASRR188 pKa = 11.84VARR191 pKa = 11.84PAPGGAVEE199 pKa = 4.25PRR201 pKa = 11.84ARR203 pKa = 11.84PARR206 pKa = 11.84RR207 pKa = 11.84RR208 pKa = 11.84PRR210 pKa = 11.84PPPSPRR216 pKa = 11.84APLRR220 pKa = 11.84SPNRR224 pKa = 11.84TVHH227 pKa = 6.04RR228 pKa = 11.84MLRR231 pKa = 11.84RR232 pKa = 11.84RR233 pKa = 11.84RR234 pKa = 11.84PTPGQMLQGAPRR246 pKa = 11.84PQRR249 pKa = 11.84ALQTQTALLLQRR261 pKa = 11.84SLRR264 pKa = 11.84PHH266 pKa = 6.07ARR268 pKa = 11.84AGRR271 pKa = 11.84GG272 pKa = 3.2
MM1 pKa = 7.96PSPLLRR7 pKa = 11.84LARR10 pKa = 11.84SASRR14 pKa = 11.84QHH16 pKa = 5.82STSSPSSPRR25 pKa = 11.84LRR27 pKa = 11.84PSPPSPPSVAVRR39 pKa = 11.84RR40 pKa = 11.84PPDD43 pKa = 4.07GPPPAPQFSTPLCPTRR59 pKa = 11.84SSPPPQRR66 pKa = 11.84RR67 pKa = 11.84PALPRR72 pKa = 11.84PTRR75 pKa = 11.84SGPRR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84AAAGHH86 pKa = 5.53EE87 pKa = 4.02RR88 pKa = 11.84RR89 pKa = 11.84ARR91 pKa = 11.84RR92 pKa = 11.84PGTPRR97 pKa = 11.84RR98 pKa = 11.84LTTTRR103 pKa = 11.84LPRR106 pKa = 11.84STWSPLSRR114 pKa = 11.84RR115 pKa = 11.84SHH117 pKa = 6.5HH118 pKa = 6.28GRR120 pKa = 11.84LPTTPLHH127 pKa = 6.63SPLAHH132 pKa = 6.98AGHH135 pKa = 7.32AVQAARR141 pKa = 11.84RR142 pKa = 11.84ARR144 pKa = 11.84PPSRR148 pKa = 11.84RR149 pKa = 11.84LHH151 pKa = 6.42RR152 pKa = 11.84AFQRR156 pKa = 11.84HH157 pKa = 5.07RR158 pKa = 11.84ALRR161 pKa = 11.84WLLLSRR167 pKa = 11.84LSSSSRR173 pKa = 11.84LSRR176 pKa = 11.84RR177 pKa = 11.84RR178 pKa = 11.84PLSRR182 pKa = 11.84RR183 pKa = 11.84SPASRR188 pKa = 11.84VARR191 pKa = 11.84PAPGGAVEE199 pKa = 4.25PRR201 pKa = 11.84ARR203 pKa = 11.84PARR206 pKa = 11.84RR207 pKa = 11.84RR208 pKa = 11.84PRR210 pKa = 11.84PPPSPRR216 pKa = 11.84APLRR220 pKa = 11.84SPNRR224 pKa = 11.84TVHH227 pKa = 6.04RR228 pKa = 11.84MLRR231 pKa = 11.84RR232 pKa = 11.84RR233 pKa = 11.84RR234 pKa = 11.84PTPGQMLQGAPRR246 pKa = 11.84PQRR249 pKa = 11.84ALQTQTALLLQRR261 pKa = 11.84SLRR264 pKa = 11.84PHH266 pKa = 6.07ARR268 pKa = 11.84AGRR271 pKa = 11.84GG272 pKa = 3.2
Molecular weight: 30.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1047096 |
30 |
2351 |
336.9 |
35.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.362 ± 0.064 | 0.625 ± 0.011 |
6.428 ± 0.038 | 5.43 ± 0.036 |
2.699 ± 0.028 | 8.915 ± 0.037 |
2.121 ± 0.023 | 3.732 ± 0.035 |
1.681 ± 0.032 | 10.183 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.769 ± 0.019 | 1.709 ± 0.024 |
5.499 ± 0.033 | 2.739 ± 0.024 |
7.15 ± 0.046 | 6.024 ± 0.034 |
6.789 ± 0.044 | 9.859 ± 0.048 |
1.419 ± 0.018 | 1.868 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
