
bacterium HR19
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 7.21
Get precalculated fractions of proteins
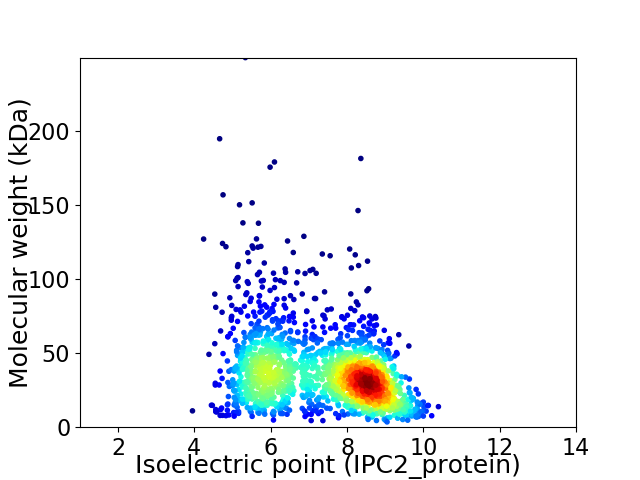
Virtual 2D-PAGE plot for 1704 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H5XST9|A0A2H5XST9_9BACT Single-stranded-DNA-specific exonuclease RecJ OS=bacterium HR19 OX=2035414 GN=recJ PE=3 SV=1
MM1 pKa = 7.87PSGFTSTSKK10 pKa = 10.66PLDD13 pKa = 4.28CNDD16 pKa = 3.84SNASIYY22 pKa = 10.42PGAPLNCNNSQDD34 pKa = 3.76NDD36 pKa = 3.5CDD38 pKa = 4.31GNVEE42 pKa = 3.8KK43 pKa = 10.46WFFQDD48 pKa = 3.66QDD50 pKa = 3.36SDD52 pKa = 3.4TWTTSISQCANTMPSGFTSTSKK74 pKa = 10.66PLDD77 pKa = 4.28CNDD80 pKa = 3.84SNASIYY86 pKa = 10.42PGAPLNCNNSQDD98 pKa = 3.76NDD100 pKa = 3.5CDD102 pKa = 4.31GNVEE106 pKa = 3.8KK107 pKa = 10.46WFFQDD112 pKa = 3.66QDD114 pKa = 3.36SDD116 pKa = 3.4TWTTSISQCANTMPSGFTSTSKK138 pKa = 10.66PLDD141 pKa = 4.28CNDD144 pKa = 3.84SNASIYY150 pKa = 10.42PGAPLNCNNSQDD162 pKa = 3.76NDD164 pKa = 3.5CDD166 pKa = 4.31GNVEE170 pKa = 3.8KK171 pKa = 10.46WFFQDD176 pKa = 3.66QDD178 pKa = 3.36SDD180 pKa = 3.42TWTTSVSQCANTMPSGFTSTSKK202 pKa = 10.68PLDD205 pKa = 4.16CNDD208 pKa = 4.15FSPLINPGQPEE219 pKa = 4.03ICNGVDD225 pKa = 3.61DD226 pKa = 4.8NCDD229 pKa = 3.23GVIDD233 pKa = 4.7DD234 pKa = 4.54VACGVPVCPTSLTATYY250 pKa = 9.27FFRR253 pKa = 11.84SSTGDD258 pKa = 3.06ISIKK262 pKa = 10.8LEE264 pKa = 4.11WNDD267 pKa = 3.23PATNEE272 pKa = 3.96TGFVVEE278 pKa = 4.65KK279 pKa = 10.31SKK281 pKa = 11.02KK282 pKa = 9.49SSFFPSITYY291 pKa = 9.33VINSPNTTTFTLDD304 pKa = 3.19FEE306 pKa = 4.79EE307 pKa = 4.78PLTRR311 pKa = 11.84FWFRR315 pKa = 11.84VWAFNSTVPCTPIAISSVFTPPGLLWAYY343 pKa = 10.2SLTSIARR350 pKa = 11.84DD351 pKa = 3.36QNNYY355 pKa = 9.09PIHH358 pKa = 6.69PKK360 pKa = 10.14IFDD363 pKa = 3.95LNGDD367 pKa = 3.82GKK369 pKa = 11.03KK370 pKa = 9.93EE371 pKa = 4.05VIMSDD376 pKa = 2.62SGGRR380 pKa = 11.84VYY382 pKa = 10.81AISTTEE388 pKa = 3.7FNLEE392 pKa = 3.48ISAIPLWIIRR402 pKa = 11.84PAGNKK407 pKa = 9.75KK408 pKa = 8.58FTTNSIAVFQGNATVIAINQQDD430 pKa = 3.64SRR432 pKa = 11.84SYY434 pKa = 10.8IIDD437 pKa = 3.25KK438 pKa = 10.54NGNVTKK444 pKa = 10.78NIALGTTPPYY454 pKa = 10.17HH455 pKa = 7.34AVITDD460 pKa = 3.27INFDD464 pKa = 4.41LLPDD468 pKa = 5.22FIVSLQNQVKK478 pKa = 10.52AFDD481 pKa = 3.92FFGNNLFTADD491 pKa = 3.74PPGAEE496 pKa = 4.41VIGTPTALIEE506 pKa = 4.33YY507 pKa = 9.28KK508 pKa = 10.78GKK510 pKa = 6.91TTTYY514 pKa = 10.66VVGTNTRR521 pKa = 11.84KK522 pKa = 9.39IRR524 pKa = 11.84FYY526 pKa = 10.81EE527 pKa = 4.24GNNQISVIDD536 pKa = 3.85LTLQPFSGGVPSFISTFKK554 pKa = 10.74KK555 pKa = 10.18GALSRR560 pKa = 11.84AVVGTDD566 pKa = 2.78TDD568 pKa = 3.49IFITNLIKK576 pKa = 11.06NPLLTFNTNAQVIATPLFFDD596 pKa = 4.68FSGDD600 pKa = 3.5GITDD604 pKa = 3.45VAITSGNFLRR614 pKa = 11.84IINGATGLEE623 pKa = 3.75ICSFNLGANTDD634 pKa = 3.73SSPVLLPTTPVQILVGANNGILYY657 pKa = 10.01SISPSCTQNWQYY669 pKa = 11.55NAGLGFQIKK678 pKa = 8.4STPLSANLDD687 pKa = 3.97SNSSIEE693 pKa = 4.34IIFTATSATSGKK705 pKa = 9.92IVILDD710 pKa = 3.46SSGNLIKK717 pKa = 10.67EE718 pKa = 3.99FSDD721 pKa = 3.75LVNIGPTVSSPRR733 pKa = 11.84AGDD736 pKa = 3.13IDD738 pKa = 4.43GNGVTDD744 pKa = 3.66VVIGGSKK751 pKa = 10.59VIGTQGGIYY760 pKa = 9.95IVTCGTDD767 pKa = 3.2CSSANPKK774 pKa = 10.12KK775 pKa = 10.27FSKK778 pKa = 10.7SGWSEE783 pKa = 3.65IKK785 pKa = 7.75TTPLLYY791 pKa = 10.61DD792 pKa = 4.4LNSDD796 pKa = 3.97GYY798 pKa = 11.28LDD800 pKa = 4.14IVVADD805 pKa = 4.34TGNNMYY811 pKa = 10.26AISGTSINYY820 pKa = 9.58SEE822 pKa = 5.93PYY824 pKa = 10.72LLWQDD829 pKa = 3.37NLTKK833 pKa = 10.14IGEE836 pKa = 4.38TPPGVVFPSSPTVFLKK852 pKa = 11.16GSIPYY857 pKa = 9.57IAIGSNGQGIYY868 pKa = 10.09IIRR871 pKa = 11.84GNDD874 pKa = 2.99GSAQNIGICNKK885 pKa = 8.8TFSSPATFDD894 pKa = 3.48YY895 pKa = 11.13NRR897 pKa = 11.84DD898 pKa = 3.58GIADD902 pKa = 4.03LVFGCDD908 pKa = 2.85NKK910 pKa = 10.35FVYY913 pKa = 10.0IISGSDD919 pKa = 3.09FSVLRR924 pKa = 11.84KK925 pKa = 9.78EE926 pKa = 4.14KK927 pKa = 10.95VGDD930 pKa = 3.75KK931 pKa = 10.85VRR933 pKa = 11.84GSPKK937 pKa = 9.93IYY939 pKa = 10.6DD940 pKa = 3.43FDD942 pKa = 6.29GDD944 pKa = 4.12GVQDD948 pKa = 3.58IGIGSDD954 pKa = 3.38DD955 pKa = 3.64KK956 pKa = 11.53NIYY959 pKa = 8.93VFNGEE964 pKa = 4.03RR965 pKa = 11.84TFICSFPTPVQNQPAIFRR983 pKa = 11.84DD984 pKa = 3.8KK985 pKa = 10.78IIFSDD990 pKa = 3.7SSNLYY995 pKa = 9.71VLNSDD1000 pKa = 3.13NCIAVAQKK1008 pKa = 10.94NGVSLSPSSSCGVFYY1023 pKa = 9.12EE1024 pKa = 4.37TAGGKK1029 pKa = 9.9EE1030 pKa = 3.57ILICGDD1036 pKa = 3.43EE1037 pKa = 4.46TNGEE1041 pKa = 4.25ILVMDD1046 pKa = 4.62SALNDD1051 pKa = 3.18IFPFPFTFASATFFSSPAIDD1071 pKa = 5.97DD1072 pKa = 3.54IDD1074 pKa = 4.9GDD1076 pKa = 3.95SSLEE1080 pKa = 3.93FVISSQDD1087 pKa = 3.0MVFAYY1092 pKa = 10.53RR1093 pKa = 11.84LTNSEE1098 pKa = 4.81FEE1100 pKa = 4.3RR1101 pKa = 11.84LWGEE1105 pKa = 4.39FSHH1108 pKa = 7.36DD1109 pKa = 3.61RR1110 pKa = 11.84YY1111 pKa = 10.9NSSFKK1116 pKa = 9.52EE1117 pKa = 4.01QKK1119 pKa = 9.28EE1120 pKa = 4.14VSYY1123 pKa = 10.54LSPNISVSKK1132 pKa = 10.56IKK1134 pKa = 10.97VPDD1137 pKa = 3.47LHH1139 pKa = 7.13EE1140 pKa = 4.79SEE1142 pKa = 5.46SKK1144 pKa = 10.88GCSTSMPYY1152 pKa = 10.29FSTMIITLSYY1162 pKa = 10.66LIFRR1166 pKa = 11.84RR1167 pKa = 11.84RR1168 pKa = 11.84KK1169 pKa = 9.11NLL1171 pKa = 3.25
MM1 pKa = 7.87PSGFTSTSKK10 pKa = 10.66PLDD13 pKa = 4.28CNDD16 pKa = 3.84SNASIYY22 pKa = 10.42PGAPLNCNNSQDD34 pKa = 3.76NDD36 pKa = 3.5CDD38 pKa = 4.31GNVEE42 pKa = 3.8KK43 pKa = 10.46WFFQDD48 pKa = 3.66QDD50 pKa = 3.36SDD52 pKa = 3.4TWTTSISQCANTMPSGFTSTSKK74 pKa = 10.66PLDD77 pKa = 4.28CNDD80 pKa = 3.84SNASIYY86 pKa = 10.42PGAPLNCNNSQDD98 pKa = 3.76NDD100 pKa = 3.5CDD102 pKa = 4.31GNVEE106 pKa = 3.8KK107 pKa = 10.46WFFQDD112 pKa = 3.66QDD114 pKa = 3.36SDD116 pKa = 3.4TWTTSISQCANTMPSGFTSTSKK138 pKa = 10.66PLDD141 pKa = 4.28CNDD144 pKa = 3.84SNASIYY150 pKa = 10.42PGAPLNCNNSQDD162 pKa = 3.76NDD164 pKa = 3.5CDD166 pKa = 4.31GNVEE170 pKa = 3.8KK171 pKa = 10.46WFFQDD176 pKa = 3.66QDD178 pKa = 3.36SDD180 pKa = 3.42TWTTSVSQCANTMPSGFTSTSKK202 pKa = 10.68PLDD205 pKa = 4.16CNDD208 pKa = 4.15FSPLINPGQPEE219 pKa = 4.03ICNGVDD225 pKa = 3.61DD226 pKa = 4.8NCDD229 pKa = 3.23GVIDD233 pKa = 4.7DD234 pKa = 4.54VACGVPVCPTSLTATYY250 pKa = 9.27FFRR253 pKa = 11.84SSTGDD258 pKa = 3.06ISIKK262 pKa = 10.8LEE264 pKa = 4.11WNDD267 pKa = 3.23PATNEE272 pKa = 3.96TGFVVEE278 pKa = 4.65KK279 pKa = 10.31SKK281 pKa = 11.02KK282 pKa = 9.49SSFFPSITYY291 pKa = 9.33VINSPNTTTFTLDD304 pKa = 3.19FEE306 pKa = 4.79EE307 pKa = 4.78PLTRR311 pKa = 11.84FWFRR315 pKa = 11.84VWAFNSTVPCTPIAISSVFTPPGLLWAYY343 pKa = 10.2SLTSIARR350 pKa = 11.84DD351 pKa = 3.36QNNYY355 pKa = 9.09PIHH358 pKa = 6.69PKK360 pKa = 10.14IFDD363 pKa = 3.95LNGDD367 pKa = 3.82GKK369 pKa = 11.03KK370 pKa = 9.93EE371 pKa = 4.05VIMSDD376 pKa = 2.62SGGRR380 pKa = 11.84VYY382 pKa = 10.81AISTTEE388 pKa = 3.7FNLEE392 pKa = 3.48ISAIPLWIIRR402 pKa = 11.84PAGNKK407 pKa = 9.75KK408 pKa = 8.58FTTNSIAVFQGNATVIAINQQDD430 pKa = 3.64SRR432 pKa = 11.84SYY434 pKa = 10.8IIDD437 pKa = 3.25KK438 pKa = 10.54NGNVTKK444 pKa = 10.78NIALGTTPPYY454 pKa = 10.17HH455 pKa = 7.34AVITDD460 pKa = 3.27INFDD464 pKa = 4.41LLPDD468 pKa = 5.22FIVSLQNQVKK478 pKa = 10.52AFDD481 pKa = 3.92FFGNNLFTADD491 pKa = 3.74PPGAEE496 pKa = 4.41VIGTPTALIEE506 pKa = 4.33YY507 pKa = 9.28KK508 pKa = 10.78GKK510 pKa = 6.91TTTYY514 pKa = 10.66VVGTNTRR521 pKa = 11.84KK522 pKa = 9.39IRR524 pKa = 11.84FYY526 pKa = 10.81EE527 pKa = 4.24GNNQISVIDD536 pKa = 3.85LTLQPFSGGVPSFISTFKK554 pKa = 10.74KK555 pKa = 10.18GALSRR560 pKa = 11.84AVVGTDD566 pKa = 2.78TDD568 pKa = 3.49IFITNLIKK576 pKa = 11.06NPLLTFNTNAQVIATPLFFDD596 pKa = 4.68FSGDD600 pKa = 3.5GITDD604 pKa = 3.45VAITSGNFLRR614 pKa = 11.84IINGATGLEE623 pKa = 3.75ICSFNLGANTDD634 pKa = 3.73SSPVLLPTTPVQILVGANNGILYY657 pKa = 10.01SISPSCTQNWQYY669 pKa = 11.55NAGLGFQIKK678 pKa = 8.4STPLSANLDD687 pKa = 3.97SNSSIEE693 pKa = 4.34IIFTATSATSGKK705 pKa = 9.92IVILDD710 pKa = 3.46SSGNLIKK717 pKa = 10.67EE718 pKa = 3.99FSDD721 pKa = 3.75LVNIGPTVSSPRR733 pKa = 11.84AGDD736 pKa = 3.13IDD738 pKa = 4.43GNGVTDD744 pKa = 3.66VVIGGSKK751 pKa = 10.59VIGTQGGIYY760 pKa = 9.95IVTCGTDD767 pKa = 3.2CSSANPKK774 pKa = 10.12KK775 pKa = 10.27FSKK778 pKa = 10.7SGWSEE783 pKa = 3.65IKK785 pKa = 7.75TTPLLYY791 pKa = 10.61DD792 pKa = 4.4LNSDD796 pKa = 3.97GYY798 pKa = 11.28LDD800 pKa = 4.14IVVADD805 pKa = 4.34TGNNMYY811 pKa = 10.26AISGTSINYY820 pKa = 9.58SEE822 pKa = 5.93PYY824 pKa = 10.72LLWQDD829 pKa = 3.37NLTKK833 pKa = 10.14IGEE836 pKa = 4.38TPPGVVFPSSPTVFLKK852 pKa = 11.16GSIPYY857 pKa = 9.57IAIGSNGQGIYY868 pKa = 10.09IIRR871 pKa = 11.84GNDD874 pKa = 2.99GSAQNIGICNKK885 pKa = 8.8TFSSPATFDD894 pKa = 3.48YY895 pKa = 11.13NRR897 pKa = 11.84DD898 pKa = 3.58GIADD902 pKa = 4.03LVFGCDD908 pKa = 2.85NKK910 pKa = 10.35FVYY913 pKa = 10.0IISGSDD919 pKa = 3.09FSVLRR924 pKa = 11.84KK925 pKa = 9.78EE926 pKa = 4.14KK927 pKa = 10.95VGDD930 pKa = 3.75KK931 pKa = 10.85VRR933 pKa = 11.84GSPKK937 pKa = 9.93IYY939 pKa = 10.6DD940 pKa = 3.43FDD942 pKa = 6.29GDD944 pKa = 4.12GVQDD948 pKa = 3.58IGIGSDD954 pKa = 3.38DD955 pKa = 3.64KK956 pKa = 11.53NIYY959 pKa = 8.93VFNGEE964 pKa = 4.03RR965 pKa = 11.84TFICSFPTPVQNQPAIFRR983 pKa = 11.84DD984 pKa = 3.8KK985 pKa = 10.78IIFSDD990 pKa = 3.7SSNLYY995 pKa = 9.71VLNSDD1000 pKa = 3.13NCIAVAQKK1008 pKa = 10.94NGVSLSPSSSCGVFYY1023 pKa = 9.12EE1024 pKa = 4.37TAGGKK1029 pKa = 9.9EE1030 pKa = 3.57ILICGDD1036 pKa = 3.43EE1037 pKa = 4.46TNGEE1041 pKa = 4.25ILVMDD1046 pKa = 4.62SALNDD1051 pKa = 3.18IFPFPFTFASATFFSSPAIDD1071 pKa = 5.97DD1072 pKa = 3.54IDD1074 pKa = 4.9GDD1076 pKa = 3.95SSLEE1080 pKa = 3.93FVISSQDD1087 pKa = 3.0MVFAYY1092 pKa = 10.53RR1093 pKa = 11.84LTNSEE1098 pKa = 4.81FEE1100 pKa = 4.3RR1101 pKa = 11.84LWGEE1105 pKa = 4.39FSHH1108 pKa = 7.36DD1109 pKa = 3.61RR1110 pKa = 11.84YY1111 pKa = 10.9NSSFKK1116 pKa = 9.52EE1117 pKa = 4.01QKK1119 pKa = 9.28EE1120 pKa = 4.14VSYY1123 pKa = 10.54LSPNISVSKK1132 pKa = 10.56IKK1134 pKa = 10.97VPDD1137 pKa = 3.47LHH1139 pKa = 7.13EE1140 pKa = 4.79SEE1142 pKa = 5.46SKK1144 pKa = 10.88GCSTSMPYY1152 pKa = 10.29FSTMIITLSYY1162 pKa = 10.66LIFRR1166 pKa = 11.84RR1167 pKa = 11.84RR1168 pKa = 11.84KK1169 pKa = 9.11NLL1171 pKa = 3.25
Molecular weight: 126.98 kDa
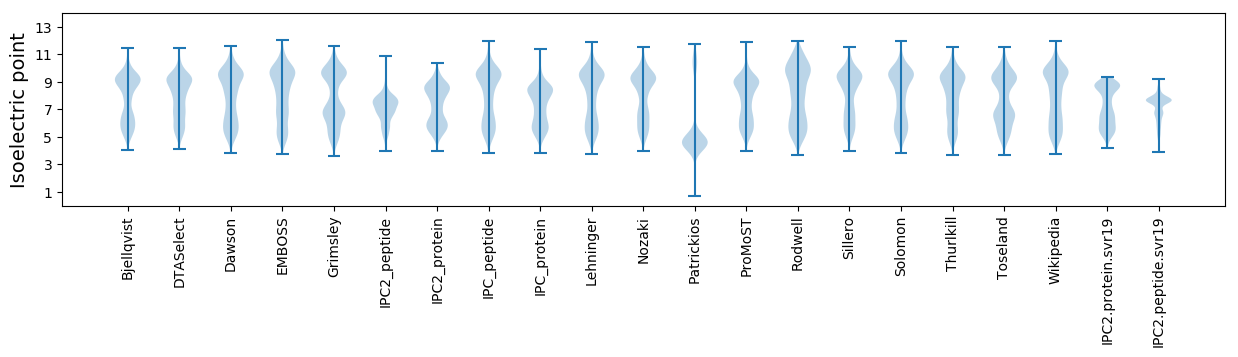
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H5XP78|A0A2H5XP78_9BACT Alanine racemase OS=bacterium HR19 OX=2035414 GN=dadX PE=3 SV=1
MM1 pKa = 7.87DD2 pKa = 3.64VFIRR6 pKa = 11.84RR7 pKa = 11.84ILANTLDD14 pKa = 3.91RR15 pKa = 11.84SVIIYY20 pKa = 10.16ISALISLVGKK30 pKa = 10.47INFLGFILMFLFFDD44 pKa = 4.51FTTSVMFRR52 pKa = 11.84WAFEE56 pKa = 4.04RR57 pKa = 11.84TPGDD61 pKa = 3.24IIFGIRR67 pKa = 11.84ISKK70 pKa = 9.52KK71 pKa = 8.27KK72 pKa = 10.08HH73 pKa = 4.49GAIHH77 pKa = 6.91LKK79 pKa = 10.48LALRR83 pKa = 11.84WIMGFISPLTGFLLHH98 pKa = 6.93IPAGNWRR105 pKa = 11.84SLCDD109 pKa = 3.59TLAGIQFEE117 pKa = 4.52RR118 pKa = 11.84VV119 pKa = 2.99
MM1 pKa = 7.87DD2 pKa = 3.64VFIRR6 pKa = 11.84RR7 pKa = 11.84ILANTLDD14 pKa = 3.91RR15 pKa = 11.84SVIIYY20 pKa = 10.16ISALISLVGKK30 pKa = 10.47INFLGFILMFLFFDD44 pKa = 4.51FTTSVMFRR52 pKa = 11.84WAFEE56 pKa = 4.04RR57 pKa = 11.84TPGDD61 pKa = 3.24IIFGIRR67 pKa = 11.84ISKK70 pKa = 9.52KK71 pKa = 8.27KK72 pKa = 10.08HH73 pKa = 4.49GAIHH77 pKa = 6.91LKK79 pKa = 10.48LALRR83 pKa = 11.84WIMGFISPLTGFLLHH98 pKa = 6.93IPAGNWRR105 pKa = 11.84SLCDD109 pKa = 3.59TLAGIQFEE117 pKa = 4.52RR118 pKa = 11.84VV119 pKa = 2.99
Molecular weight: 13.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
576888 |
31 |
2244 |
338.5 |
38.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.267 ± 0.047 | 0.963 ± 0.02 |
4.895 ± 0.037 | 8.372 ± 0.068 |
6.482 ± 0.07 | 6.559 ± 0.047 |
1.281 ± 0.019 | 9.587 ± 0.055 |
9.4 ± 0.069 | 8.873 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.852 ± 0.024 | 3.841 ± 0.038 |
3.954 ± 0.034 | 2.294 ± 0.026 |
4.58 ± 0.038 | 7.164 ± 0.063 |
3.848 ± 0.038 | 6.464 ± 0.045 |
0.954 ± 0.019 | 3.37 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
