
Sulfolobus sp. SCGC AB-777_L09
Taxonomy: cellular organisms; Archaea; TACK group; Crenarchaeota; Thermoprotei; Sulfolobales; Sulfolobaceae; Sulfolobus; unclassified Sulfolobus
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
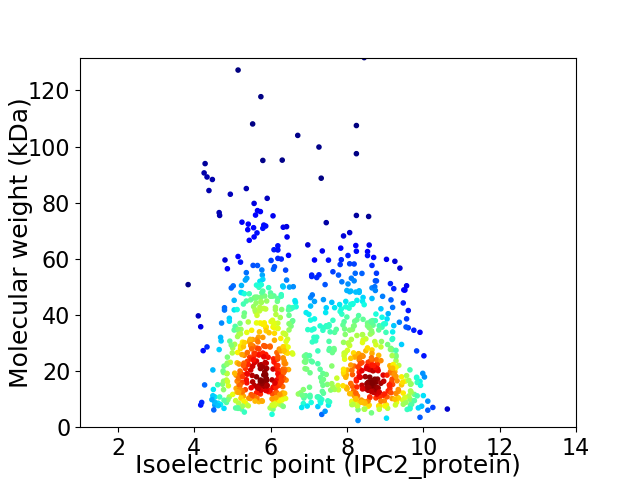
Virtual 2D-PAGE plot for 930 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T9WQU8|A0A2T9WQU8_9CREN Amidohydro_3 domain-containing protein OS=Sulfolobus sp. SCGC AB-777_L09 OX=1987492 GN=DDW09_02820 PE=4 SV=1
MM1 pKa = 7.36PPNWVFNYY9 pKa = 9.61GALANRR15 pKa = 11.84VTYY18 pKa = 9.98VVNPQIPIIVTYY30 pKa = 8.14MVPNPNGQGGPQGDD44 pKa = 3.97VQAVIIGNQVFFEE57 pKa = 4.29PSVNDD62 pKa = 4.55FSSCYY67 pKa = 10.26NGNDD71 pKa = 3.32CSALQAEE78 pKa = 4.71GLDD81 pKa = 4.1LSFLGYY87 pKa = 9.87PYY89 pKa = 10.47FITGGGCISGSGSCPPTPNPSGVEE113 pKa = 3.94TCNLIINKK121 pKa = 7.18EE122 pKa = 4.26TGEE125 pKa = 3.95ITQLFGGALSNTLSVMADD143 pKa = 3.37TNNGYY148 pKa = 10.48LIIVTSTNSTSADD161 pKa = 3.98FIIAIPFSSITSILAGQIPPDD182 pKa = 4.06AKK184 pKa = 9.65WVIVSTSDD192 pKa = 4.15GSSLYY197 pKa = 10.97ANQAEE202 pKa = 4.79LIDD205 pKa = 3.88GNIYY209 pKa = 10.28FVSNTGNGVEE219 pKa = 4.29VFKK222 pKa = 11.14FPLSEE227 pKa = 4.34VYY229 pKa = 10.48NSQCTGIPQGYY240 pKa = 8.93SSCTYY245 pKa = 8.9TVNPLTSISQPVANSGGNILTDD267 pKa = 4.79AIKK270 pKa = 10.45MNGVSYY276 pKa = 10.39LIIFYY281 pKa = 9.92PSNGNFTVILYY292 pKa = 8.0NTEE295 pKa = 4.12DD296 pKa = 3.2NTYY299 pKa = 10.61SINTFGQNIEE309 pKa = 4.16PQFITTGIAIYY320 pKa = 10.22NHH322 pKa = 6.38IVLATAQQLDD332 pKa = 4.67GTWLLYY338 pKa = 10.63AYY340 pKa = 10.21DD341 pKa = 4.18PFNGKK346 pKa = 9.18VDD348 pKa = 3.44STTIPNVYY356 pKa = 9.82QAVPTDD362 pKa = 3.17NGYY365 pKa = 10.51VVTLSNTLSSNSQVMISIYY384 pKa = 10.35KK385 pKa = 9.76VLFTISPKK393 pKa = 9.82FQNVSITKK401 pKa = 8.71TLNQVTVQGTLIDD414 pKa = 4.16EE415 pKa = 4.67NSGKK419 pKa = 9.45PLPNYY424 pKa = 9.71PVMLVAVDD432 pKa = 3.95GVEE435 pKa = 4.26VNTVTDD441 pKa = 3.82FTVLATGTTDD451 pKa = 3.16SNGNFTLTTSDD462 pKa = 3.64TSHH465 pKa = 6.36SFYY468 pKa = 11.21GVVYY472 pKa = 10.18YY473 pKa = 9.48PSS475 pKa = 3.18
MM1 pKa = 7.36PPNWVFNYY9 pKa = 9.61GALANRR15 pKa = 11.84VTYY18 pKa = 9.98VVNPQIPIIVTYY30 pKa = 8.14MVPNPNGQGGPQGDD44 pKa = 3.97VQAVIIGNQVFFEE57 pKa = 4.29PSVNDD62 pKa = 4.55FSSCYY67 pKa = 10.26NGNDD71 pKa = 3.32CSALQAEE78 pKa = 4.71GLDD81 pKa = 4.1LSFLGYY87 pKa = 9.87PYY89 pKa = 10.47FITGGGCISGSGSCPPTPNPSGVEE113 pKa = 3.94TCNLIINKK121 pKa = 7.18EE122 pKa = 4.26TGEE125 pKa = 3.95ITQLFGGALSNTLSVMADD143 pKa = 3.37TNNGYY148 pKa = 10.48LIIVTSTNSTSADD161 pKa = 3.98FIIAIPFSSITSILAGQIPPDD182 pKa = 4.06AKK184 pKa = 9.65WVIVSTSDD192 pKa = 4.15GSSLYY197 pKa = 10.97ANQAEE202 pKa = 4.79LIDD205 pKa = 3.88GNIYY209 pKa = 10.28FVSNTGNGVEE219 pKa = 4.29VFKK222 pKa = 11.14FPLSEE227 pKa = 4.34VYY229 pKa = 10.48NSQCTGIPQGYY240 pKa = 8.93SSCTYY245 pKa = 8.9TVNPLTSISQPVANSGGNILTDD267 pKa = 4.79AIKK270 pKa = 10.45MNGVSYY276 pKa = 10.39LIIFYY281 pKa = 9.92PSNGNFTVILYY292 pKa = 8.0NTEE295 pKa = 4.12DD296 pKa = 3.2NTYY299 pKa = 10.61SINTFGQNIEE309 pKa = 4.16PQFITTGIAIYY320 pKa = 10.22NHH322 pKa = 6.38IVLATAQQLDD332 pKa = 4.67GTWLLYY338 pKa = 10.63AYY340 pKa = 10.21DD341 pKa = 4.18PFNGKK346 pKa = 9.18VDD348 pKa = 3.44STTIPNVYY356 pKa = 9.82QAVPTDD362 pKa = 3.17NGYY365 pKa = 10.51VVTLSNTLSSNSQVMISIYY384 pKa = 10.35KK385 pKa = 9.76VLFTISPKK393 pKa = 9.82FQNVSITKK401 pKa = 8.71TLNQVTVQGTLIDD414 pKa = 4.16EE415 pKa = 4.67NSGKK419 pKa = 9.45PLPNYY424 pKa = 9.71PVMLVAVDD432 pKa = 3.95GVEE435 pKa = 4.26VNTVTDD441 pKa = 3.82FTVLATGTTDD451 pKa = 3.16SNGNFTLTTSDD462 pKa = 3.64TSHH465 pKa = 6.36SFYY468 pKa = 11.21GVVYY472 pKa = 10.18YY473 pKa = 9.48PSS475 pKa = 3.18
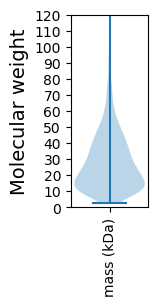
Molecular weight: 50.8 kDa
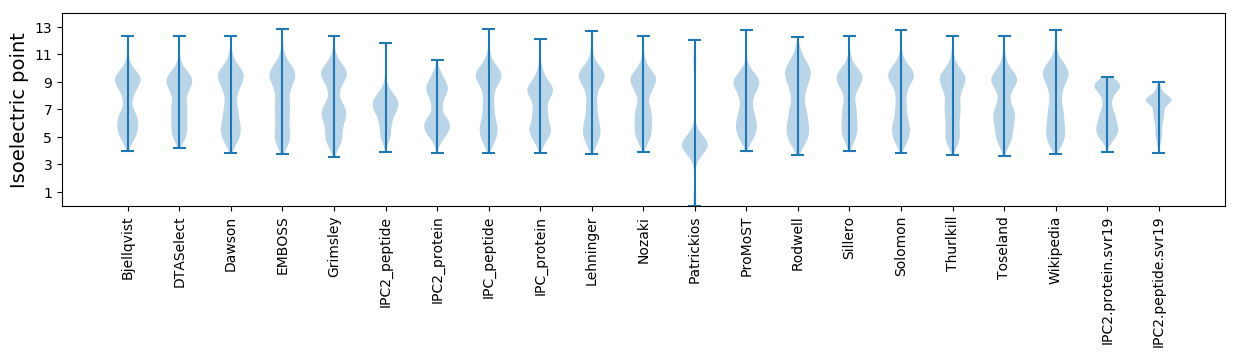
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T9WMY4|A0A2T9WMY4_9CREN NAD-dependent dehydratase OS=Sulfolobus sp. SCGC AB-777_L09 OX=1987492 GN=DDW09_04750 PE=4 SV=1
MM1 pKa = 7.05GRR3 pKa = 11.84KK4 pKa = 8.53PVFRR8 pKa = 11.84QDD10 pKa = 3.35VSCPSCGSHH19 pKa = 6.69HH20 pKa = 5.61VVKK23 pKa = 10.62CGRR26 pKa = 11.84PLGRR30 pKa = 11.84QRR32 pKa = 11.84YY33 pKa = 8.26LCRR36 pKa = 11.84DD37 pKa = 3.34CGKK40 pKa = 10.41YY41 pKa = 10.27FLGDD45 pKa = 3.29ASYY48 pKa = 10.17HH49 pKa = 4.74HH50 pKa = 7.29HH51 pKa = 5.77SRR53 pKa = 11.84KK54 pKa = 9.55LRR56 pKa = 11.84EE57 pKa = 3.83EE58 pKa = 4.14ALRR61 pKa = 11.84MYY63 pKa = 10.85ANGMSMRR70 pKa = 11.84AISRR74 pKa = 11.84VLNVPLGTVFTT85 pKa = 4.86
MM1 pKa = 7.05GRR3 pKa = 11.84KK4 pKa = 8.53PVFRR8 pKa = 11.84QDD10 pKa = 3.35VSCPSCGSHH19 pKa = 6.69HH20 pKa = 5.61VVKK23 pKa = 10.62CGRR26 pKa = 11.84PLGRR30 pKa = 11.84QRR32 pKa = 11.84YY33 pKa = 8.26LCRR36 pKa = 11.84DD37 pKa = 3.34CGKK40 pKa = 10.41YY41 pKa = 10.27FLGDD45 pKa = 3.29ASYY48 pKa = 10.17HH49 pKa = 4.74HH50 pKa = 7.29HH51 pKa = 5.77SRR53 pKa = 11.84KK54 pKa = 9.55LRR56 pKa = 11.84EE57 pKa = 3.83EE58 pKa = 4.14ALRR61 pKa = 11.84MYY63 pKa = 10.85ANGMSMRR70 pKa = 11.84AISRR74 pKa = 11.84VLNVPLGTVFTT85 pKa = 4.86
Molecular weight: 9.7 kDa
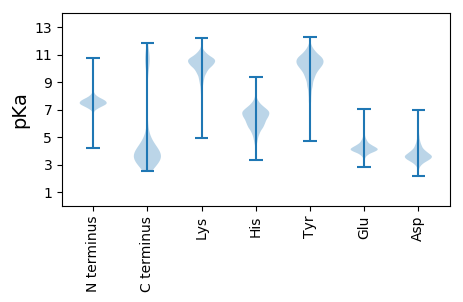
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
236844 |
21 |
1158 |
254.7 |
28.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.494 ± 0.084 | 0.643 ± 0.023 |
4.693 ± 0.065 | 7.123 ± 0.113 |
4.481 ± 0.065 | 6.603 ± 0.077 |
1.343 ± 0.03 | 9.41 ± 0.08 |
7.898 ± 0.122 | 10.244 ± 0.089 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.19 ± 0.038 | 4.677 ± 0.076 |
4.014 ± 0.063 | 2.109 ± 0.047 |
4.286 ± 0.065 | 6.506 ± 0.072 |
4.777 ± 0.066 | 7.712 ± 0.066 |
0.991 ± 0.03 | 4.805 ± 0.067 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
