
Methylophilales bacterium MBRS-H7
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; unclassified Nitrosomonadales; OM43 clade
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
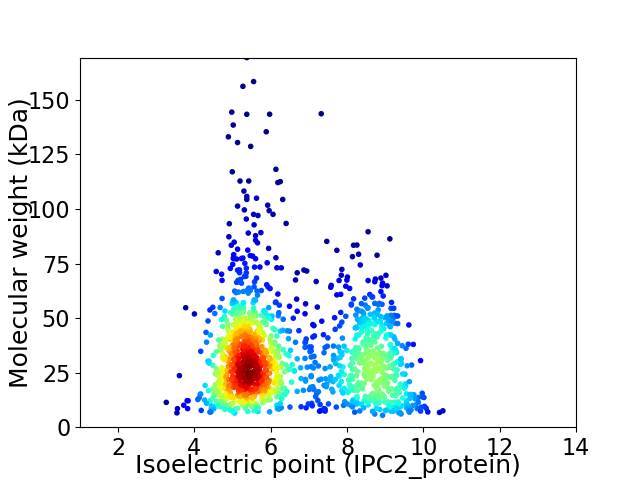
Virtual 2D-PAGE plot for 1317 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H4JA03|A0A0H4JA03_9PROT Uncharacterized protein OS=Methylophilales bacterium MBRS-H7 OX=1623450 GN=VI33_00670 PE=4 SV=1
MM1 pKa = 7.37TYY3 pKa = 10.71VVTEE7 pKa = 3.67NCIQCKK13 pKa = 8.64YY14 pKa = 8.09TDD16 pKa = 4.36CVDD19 pKa = 3.61VCPVDD24 pKa = 4.53CFVEE28 pKa = 4.96GPNFLAIDD36 pKa = 3.86PEE38 pKa = 4.21EE39 pKa = 5.95CIDD42 pKa = 3.78CTLCVAEE49 pKa = 5.38CPAEE53 pKa = 4.5AIFAEE58 pKa = 4.6DD59 pKa = 5.49DD60 pKa = 3.81VPEE63 pKa = 4.55DD64 pKa = 3.23QLDD67 pKa = 4.22FIEE70 pKa = 5.16INAKK74 pKa = 9.12LAKK77 pKa = 8.49VWPIISEE84 pKa = 4.3RR85 pKa = 11.84KK86 pKa = 10.12DD87 pKa = 3.59PLPDD91 pKa = 4.26ADD93 pKa = 4.16SFNGKK98 pKa = 8.39EE99 pKa = 4.46GKK101 pKa = 9.77RR102 pKa = 11.84EE103 pKa = 3.94LLIEE107 pKa = 4.14EE108 pKa = 4.43
MM1 pKa = 7.37TYY3 pKa = 10.71VVTEE7 pKa = 3.67NCIQCKK13 pKa = 8.64YY14 pKa = 8.09TDD16 pKa = 4.36CVDD19 pKa = 3.61VCPVDD24 pKa = 4.53CFVEE28 pKa = 4.96GPNFLAIDD36 pKa = 3.86PEE38 pKa = 4.21EE39 pKa = 5.95CIDD42 pKa = 3.78CTLCVAEE49 pKa = 5.38CPAEE53 pKa = 4.5AIFAEE58 pKa = 4.6DD59 pKa = 5.49DD60 pKa = 3.81VPEE63 pKa = 4.55DD64 pKa = 3.23QLDD67 pKa = 4.22FIEE70 pKa = 5.16INAKK74 pKa = 9.12LAKK77 pKa = 8.49VWPIISEE84 pKa = 4.3RR85 pKa = 11.84KK86 pKa = 10.12DD87 pKa = 3.59PLPDD91 pKa = 4.26ADD93 pKa = 4.16SFNGKK98 pKa = 8.39EE99 pKa = 4.46GKK101 pKa = 9.77RR102 pKa = 11.84EE103 pKa = 3.94LLIEE107 pKa = 4.14EE108 pKa = 4.43
Molecular weight: 12.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H4IXL4|A0A0H4IXL4_9PROT Uncharacterized protein OS=Methylophilales bacterium MBRS-H7 OX=1623450 GN=VI33_03025 PE=4 SV=1
MM1 pKa = 6.74ATYY4 pKa = 10.4RR5 pKa = 11.84KK6 pKa = 9.82RR7 pKa = 11.84NNKK10 pKa = 6.97WQVRR14 pKa = 11.84IQRR17 pKa = 11.84RR18 pKa = 11.84NNPPISKK25 pKa = 8.44TFVYY29 pKa = 10.61KK30 pKa = 10.61EE31 pKa = 4.01DD32 pKa = 3.47AKK34 pKa = 11.11VWARR38 pKa = 11.84QQEE41 pKa = 4.1RR42 pKa = 11.84EE43 pKa = 3.74IDD45 pKa = 3.57LGIEE49 pKa = 4.14VANQDD54 pKa = 3.32IALKK58 pKa = 9.53TLLQRR63 pKa = 11.84YY64 pKa = 7.27QKK66 pKa = 10.66EE67 pKa = 3.66ILPTKK72 pKa = 10.16KK73 pKa = 9.79HH74 pKa = 5.68PQPDD78 pKa = 3.3NYY80 pKa = 10.38RR81 pKa = 11.84INKK84 pKa = 8.84ILLHH88 pKa = 6.52PIVSLKK94 pKa = 9.75MRR96 pKa = 11.84TIKK99 pKa = 10.31SQHH102 pKa = 5.74ISSFRR107 pKa = 11.84DD108 pKa = 3.28QLIKK112 pKa = 10.35EE113 pKa = 4.36LKK115 pKa = 9.63SPNTIRR121 pKa = 11.84LYY123 pKa = 10.9LAILSHH129 pKa = 7.06LFTIARR135 pKa = 11.84TEE137 pKa = 3.8WGFDD141 pKa = 3.32NLNNPVLKK149 pKa = 10.04IRR151 pKa = 11.84RR152 pKa = 11.84PRR154 pKa = 11.84LPQSRR159 pKa = 11.84DD160 pKa = 2.99TRR162 pKa = 11.84LSDD165 pKa = 4.34DD166 pKa = 4.87DD167 pKa = 3.34IHH169 pKa = 7.42RR170 pKa = 11.84ICQHH174 pKa = 5.44SQSSLLPLLIHH185 pKa = 6.85IALDD189 pKa = 3.17TAMRR193 pKa = 11.84VSEE196 pKa = 4.14IVKK199 pKa = 10.28LRR201 pKa = 11.84VNDD204 pKa = 3.7CDD206 pKa = 3.79FKK208 pKa = 11.52KK209 pKa = 10.85RR210 pKa = 11.84MITVRR215 pKa = 11.84NTKK218 pKa = 9.91NYY220 pKa = 7.94HH221 pKa = 6.09DD222 pKa = 5.07RR223 pKa = 11.84YY224 pKa = 10.59IPMTNKK230 pKa = 9.03VAKK233 pKa = 9.84ILQHH237 pKa = 3.95QTRR240 pKa = 11.84LQGEE244 pKa = 4.23RR245 pKa = 11.84LFNITSHH252 pKa = 5.8AVSVAFYY259 pKa = 9.58RR260 pKa = 11.84ACKK263 pKa = 9.76RR264 pKa = 11.84AGIPNASFHH273 pKa = 5.65TLRR276 pKa = 11.84HH277 pKa = 5.26EE278 pKa = 4.07AVSRR282 pKa = 11.84FFEE285 pKa = 4.53KK286 pKa = 10.64GLNPMEE292 pKa = 4.18VATISGHH299 pKa = 6.37QSMQVLKK306 pKa = 10.18RR307 pKa = 11.84YY308 pKa = 6.64THH310 pKa = 6.2IQTTHH315 pKa = 6.88LLGKK319 pKa = 8.87MMRR322 pKa = 3.96
MM1 pKa = 6.74ATYY4 pKa = 10.4RR5 pKa = 11.84KK6 pKa = 9.82RR7 pKa = 11.84NNKK10 pKa = 6.97WQVRR14 pKa = 11.84IQRR17 pKa = 11.84RR18 pKa = 11.84NNPPISKK25 pKa = 8.44TFVYY29 pKa = 10.61KK30 pKa = 10.61EE31 pKa = 4.01DD32 pKa = 3.47AKK34 pKa = 11.11VWARR38 pKa = 11.84QQEE41 pKa = 4.1RR42 pKa = 11.84EE43 pKa = 3.74IDD45 pKa = 3.57LGIEE49 pKa = 4.14VANQDD54 pKa = 3.32IALKK58 pKa = 9.53TLLQRR63 pKa = 11.84YY64 pKa = 7.27QKK66 pKa = 10.66EE67 pKa = 3.66ILPTKK72 pKa = 10.16KK73 pKa = 9.79HH74 pKa = 5.68PQPDD78 pKa = 3.3NYY80 pKa = 10.38RR81 pKa = 11.84INKK84 pKa = 8.84ILLHH88 pKa = 6.52PIVSLKK94 pKa = 9.75MRR96 pKa = 11.84TIKK99 pKa = 10.31SQHH102 pKa = 5.74ISSFRR107 pKa = 11.84DD108 pKa = 3.28QLIKK112 pKa = 10.35EE113 pKa = 4.36LKK115 pKa = 9.63SPNTIRR121 pKa = 11.84LYY123 pKa = 10.9LAILSHH129 pKa = 7.06LFTIARR135 pKa = 11.84TEE137 pKa = 3.8WGFDD141 pKa = 3.32NLNNPVLKK149 pKa = 10.04IRR151 pKa = 11.84RR152 pKa = 11.84PRR154 pKa = 11.84LPQSRR159 pKa = 11.84DD160 pKa = 2.99TRR162 pKa = 11.84LSDD165 pKa = 4.34DD166 pKa = 4.87DD167 pKa = 3.34IHH169 pKa = 7.42RR170 pKa = 11.84ICQHH174 pKa = 5.44SQSSLLPLLIHH185 pKa = 6.85IALDD189 pKa = 3.17TAMRR193 pKa = 11.84VSEE196 pKa = 4.14IVKK199 pKa = 10.28LRR201 pKa = 11.84VNDD204 pKa = 3.7CDD206 pKa = 3.79FKK208 pKa = 11.52KK209 pKa = 10.85RR210 pKa = 11.84MITVRR215 pKa = 11.84NTKK218 pKa = 9.91NYY220 pKa = 7.94HH221 pKa = 6.09DD222 pKa = 5.07RR223 pKa = 11.84YY224 pKa = 10.59IPMTNKK230 pKa = 9.03VAKK233 pKa = 9.84ILQHH237 pKa = 3.95QTRR240 pKa = 11.84LQGEE244 pKa = 4.23RR245 pKa = 11.84LFNITSHH252 pKa = 5.8AVSVAFYY259 pKa = 9.58RR260 pKa = 11.84ACKK263 pKa = 9.76RR264 pKa = 11.84AGIPNASFHH273 pKa = 5.65TLRR276 pKa = 11.84HH277 pKa = 5.26EE278 pKa = 4.07AVSRR282 pKa = 11.84FFEE285 pKa = 4.53KK286 pKa = 10.64GLNPMEE292 pKa = 4.18VATISGHH299 pKa = 6.37QSMQVLKK306 pKa = 10.18RR307 pKa = 11.84YY308 pKa = 6.64THH310 pKa = 6.2IQTTHH315 pKa = 6.88LLGKK319 pKa = 8.87MMRR322 pKa = 3.96
Molecular weight: 37.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
403159 |
50 |
1541 |
306.1 |
34.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.292 ± 0.069 | 0.92 ± 0.019 |
5.891 ± 0.052 | 6.286 ± 0.075 |
5.049 ± 0.066 | 6.113 ± 0.07 |
2.084 ± 0.032 | 8.967 ± 0.066 |
8.047 ± 0.077 | 9.698 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.392 ± 0.03 | 5.978 ± 0.06 |
3.502 ± 0.038 | 3.626 ± 0.036 |
3.585 ± 0.043 | 6.808 ± 0.052 |
4.709 ± 0.037 | 5.926 ± 0.056 |
0.911 ± 0.021 | 3.215 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
