
Nocardioides sp.
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides; unclassified Nocardioides
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
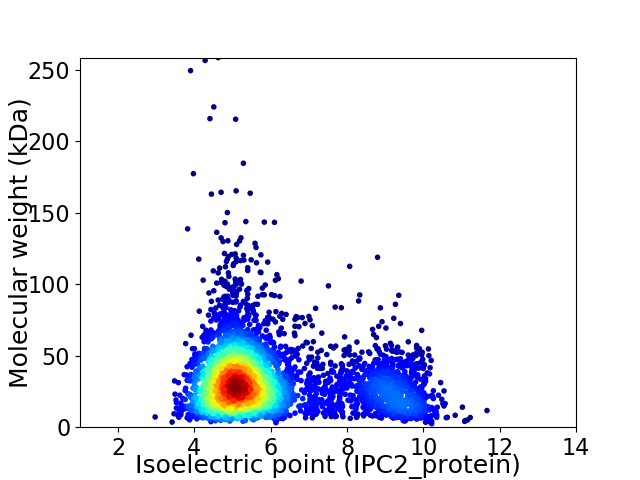
Virtual 2D-PAGE plot for 4389 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4S5AF19|A0A4S5AF19_9ACTN Biotin synthase OS=Nocardioides sp. OX=35761 GN=bioB PE=3 SV=1
MM1 pKa = 7.3NVTPFRR7 pKa = 11.84RR8 pKa = 11.84ALVPGGAALALILTACGGGAEE29 pKa = 4.51GGSGASGSVTVDD41 pKa = 3.35GSSTVYY47 pKa = 10.48PMSSAAAEE55 pKa = 4.05LLADD59 pKa = 4.09EE60 pKa = 5.03NSDD63 pKa = 3.41IEE65 pKa = 4.5VSVGFAGTGGGFEE78 pKa = 4.72KK79 pKa = 10.63FCSGDD84 pKa = 3.27TDD86 pKa = 2.98ISNASRR92 pKa = 11.84PIKK95 pKa = 10.53DD96 pKa = 4.01EE97 pKa = 3.9EE98 pKa = 4.14ATACTDD104 pKa = 3.06AGIEE108 pKa = 4.08YY109 pKa = 9.99TEE111 pKa = 4.81LLVATDD117 pKa = 3.44ALTVVVHH124 pKa = 6.89PDD126 pKa = 3.32LAVDD130 pKa = 4.3CLTTEE135 pKa = 4.03QVVEE139 pKa = 3.91LFGPNSKK146 pKa = 8.03VTKK149 pKa = 9.98WSDD152 pKa = 3.48LDD154 pKa = 3.73PSFPDD159 pKa = 3.19TKK161 pKa = 10.85VAGFIPGTDD170 pKa = 3.11SGTYY174 pKa = 10.12DD175 pKa = 3.39YY176 pKa = 10.64MAADD180 pKa = 3.71VVGDD184 pKa = 3.51EE185 pKa = 4.74SEE187 pKa = 4.16TLRR190 pKa = 11.84EE191 pKa = 4.13DD192 pKa = 3.93FEE194 pKa = 4.99SSEE197 pKa = 4.18DD198 pKa = 4.37DD199 pKa = 3.43NVLVQGVSGTEE210 pKa = 3.89GAVGFFGYY218 pKa = 9.22TYY220 pKa = 10.69FEE222 pKa = 4.47EE223 pKa = 4.63NADD226 pKa = 3.71KK227 pKa = 11.24VKK229 pKa = 10.75ALQIDD234 pKa = 4.03DD235 pKa = 4.24GQGCVAPSAEE245 pKa = 4.06TAQAGEE251 pKa = 4.43YY252 pKa = 9.36TPLARR257 pKa = 11.84PLFIYY262 pKa = 10.46VNNAKK267 pKa = 10.73YY268 pKa = 10.73NDD270 pKa = 3.64NEE272 pKa = 4.12AVKK275 pKa = 10.16AYY277 pKa = 9.39TDD279 pKa = 4.01FYY281 pKa = 10.94IEE283 pKa = 3.99NLAAIAKK290 pKa = 6.88GAKK293 pKa = 9.44YY294 pKa = 10.17IPLNDD299 pKa = 3.26EE300 pKa = 5.21DD301 pKa = 4.12YY302 pKa = 11.66AEE304 pKa = 4.4TKK306 pKa = 9.4TALEE310 pKa = 4.36GLAGG314 pKa = 3.71
MM1 pKa = 7.3NVTPFRR7 pKa = 11.84RR8 pKa = 11.84ALVPGGAALALILTACGGGAEE29 pKa = 4.51GGSGASGSVTVDD41 pKa = 3.35GSSTVYY47 pKa = 10.48PMSSAAAEE55 pKa = 4.05LLADD59 pKa = 4.09EE60 pKa = 5.03NSDD63 pKa = 3.41IEE65 pKa = 4.5VSVGFAGTGGGFEE78 pKa = 4.72KK79 pKa = 10.63FCSGDD84 pKa = 3.27TDD86 pKa = 2.98ISNASRR92 pKa = 11.84PIKK95 pKa = 10.53DD96 pKa = 4.01EE97 pKa = 3.9EE98 pKa = 4.14ATACTDD104 pKa = 3.06AGIEE108 pKa = 4.08YY109 pKa = 9.99TEE111 pKa = 4.81LLVATDD117 pKa = 3.44ALTVVVHH124 pKa = 6.89PDD126 pKa = 3.32LAVDD130 pKa = 4.3CLTTEE135 pKa = 4.03QVVEE139 pKa = 3.91LFGPNSKK146 pKa = 8.03VTKK149 pKa = 9.98WSDD152 pKa = 3.48LDD154 pKa = 3.73PSFPDD159 pKa = 3.19TKK161 pKa = 10.85VAGFIPGTDD170 pKa = 3.11SGTYY174 pKa = 10.12DD175 pKa = 3.39YY176 pKa = 10.64MAADD180 pKa = 3.71VVGDD184 pKa = 3.51EE185 pKa = 4.74SEE187 pKa = 4.16TLRR190 pKa = 11.84EE191 pKa = 4.13DD192 pKa = 3.93FEE194 pKa = 4.99SSEE197 pKa = 4.18DD198 pKa = 4.37DD199 pKa = 3.43NVLVQGVSGTEE210 pKa = 3.89GAVGFFGYY218 pKa = 9.22TYY220 pKa = 10.69FEE222 pKa = 4.47EE223 pKa = 4.63NADD226 pKa = 3.71KK227 pKa = 11.24VKK229 pKa = 10.75ALQIDD234 pKa = 4.03DD235 pKa = 4.24GQGCVAPSAEE245 pKa = 4.06TAQAGEE251 pKa = 4.43YY252 pKa = 9.36TPLARR257 pKa = 11.84PLFIYY262 pKa = 10.46VNNAKK267 pKa = 10.73YY268 pKa = 10.73NDD270 pKa = 3.64NEE272 pKa = 4.12AVKK275 pKa = 10.16AYY277 pKa = 9.39TDD279 pKa = 4.01FYY281 pKa = 10.94IEE283 pKa = 3.99NLAAIAKK290 pKa = 6.88GAKK293 pKa = 9.44YY294 pKa = 10.17IPLNDD299 pKa = 3.26EE300 pKa = 5.21DD301 pKa = 4.12YY302 pKa = 11.66AEE304 pKa = 4.4TKK306 pKa = 9.4TALEE310 pKa = 4.36GLAGG314 pKa = 3.71
Molecular weight: 32.73 kDa
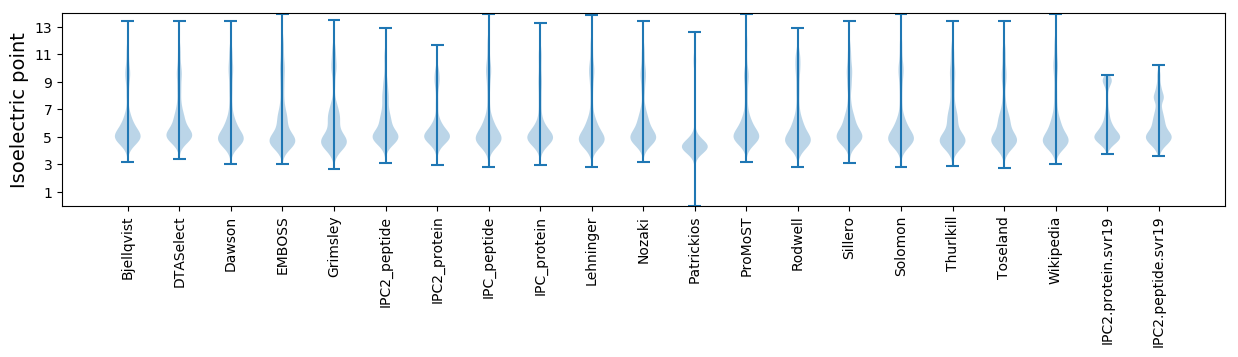
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4S4ZRU8|A0A4S4ZRU8_9ACTN SigE family RNA polymerase sigma factor OS=Nocardioides sp. OX=35761 GN=E7Z54_19300 PE=3 SV=1
RR1 pKa = 7.7RR2 pKa = 11.84PTGAGRR8 pKa = 11.84RR9 pKa = 11.84PRR11 pKa = 11.84PHH13 pKa = 7.24RR14 pKa = 11.84GGTAHH19 pKa = 7.27GAAAGRR25 pKa = 11.84APRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84GGARR34 pKa = 11.84EE35 pKa = 3.66PRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84PGRR42 pKa = 11.84RR43 pKa = 11.84ARR45 pKa = 11.84GRR47 pKa = 11.84ARR49 pKa = 11.84RR50 pKa = 11.84APGRR54 pKa = 11.84VAGARR59 pKa = 11.84GPQPP63 pKa = 3.25
RR1 pKa = 7.7RR2 pKa = 11.84PTGAGRR8 pKa = 11.84RR9 pKa = 11.84PRR11 pKa = 11.84PHH13 pKa = 7.24RR14 pKa = 11.84GGTAHH19 pKa = 7.27GAAAGRR25 pKa = 11.84APRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84GGARR34 pKa = 11.84EE35 pKa = 3.66PRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84PGRR42 pKa = 11.84RR43 pKa = 11.84ARR45 pKa = 11.84GRR47 pKa = 11.84ARR49 pKa = 11.84RR50 pKa = 11.84APGRR54 pKa = 11.84VAGARR59 pKa = 11.84GPQPP63 pKa = 3.25
Molecular weight: 6.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1381642 |
24 |
2468 |
314.8 |
33.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.922 ± 0.047 | 0.763 ± 0.01 |
6.414 ± 0.028 | 5.951 ± 0.032 |
2.818 ± 0.02 | 9.166 ± 0.031 |
2.222 ± 0.019 | 3.687 ± 0.026 |
1.976 ± 0.025 | 10.14 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.86 ± 0.013 | 1.846 ± 0.021 |
5.612 ± 0.024 | 2.785 ± 0.019 |
7.799 ± 0.041 | 5.237 ± 0.023 |
5.963 ± 0.039 | 9.297 ± 0.033 |
1.524 ± 0.016 | 2.016 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
