
Punica granatum (Pomegranate)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Lythraceae; Punica
Average proteome isoelectric point is 7.13
Get precalculated fractions of proteins
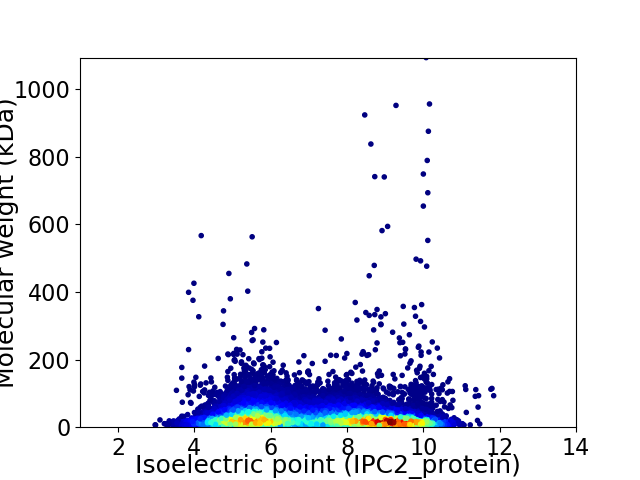
Virtual 2D-PAGE plot for 50426 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
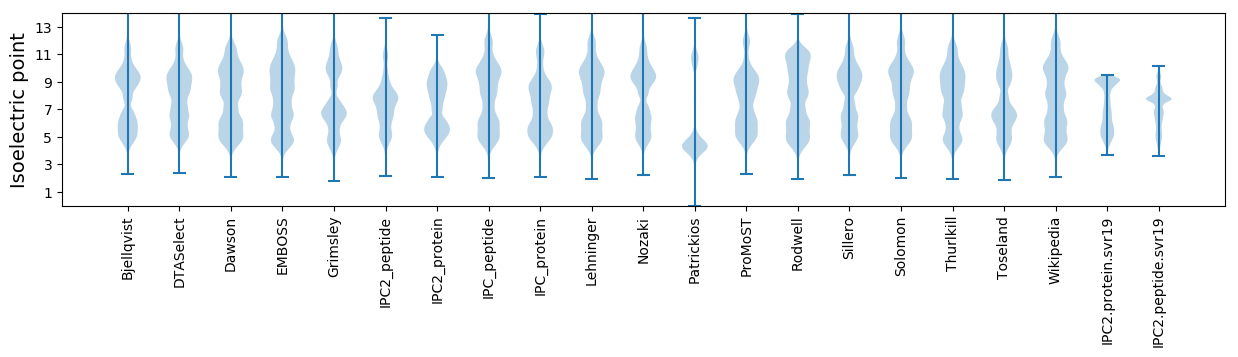
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A218XTQ1|A0A218XTQ1_PUNGR uncharacterized protein LOC116215032 OS=Punica granatum OX=22663 GN=LOC116215032 PE=4 SV=1
MM1 pKa = 7.57ASSLVLEE8 pKa = 4.63SRR10 pKa = 11.84GIANNVKK17 pKa = 9.98LHH19 pKa = 6.67EE20 pKa = 4.9DD21 pKa = 3.51FEE23 pKa = 6.56GIDD26 pKa = 3.76VEE28 pKa = 4.34VLEE31 pKa = 4.95INSSILKK38 pKa = 10.42SLLEE42 pKa = 4.39DD43 pKa = 4.11AQGDD47 pKa = 4.07DD48 pKa = 4.31QDD50 pKa = 4.28YY51 pKa = 11.17DD52 pKa = 3.44QLNDD56 pKa = 3.5VIRR59 pKa = 11.84SLEE62 pKa = 3.93AEE64 pKa = 4.23IDD66 pKa = 3.75PSSLAGHH73 pKa = 7.37DD74 pKa = 4.25LGTHH78 pKa = 5.31TKK80 pKa = 10.33VINDD84 pKa = 4.04LVAGDD89 pKa = 4.25EE90 pKa = 4.06NSTINSRR97 pKa = 11.84GRR99 pKa = 11.84DD100 pKa = 3.54GSRR103 pKa = 11.84DD104 pKa = 3.0WSEE107 pKa = 3.91YY108 pKa = 10.51SSDD111 pKa = 3.54DD112 pKa = 4.09FSWIDD117 pKa = 3.42MDD119 pKa = 4.61TVPSSPGEE127 pKa = 4.0GMDD130 pKa = 2.81WYY132 pKa = 10.36MEE134 pKa = 4.14EE135 pKa = 5.55FGDD138 pKa = 5.23EE139 pKa = 4.49MNCLTQFGCIGDD151 pKa = 3.96YY152 pKa = 10.05YY153 pKa = 10.74QYY155 pKa = 9.6PQVYY159 pKa = 9.7SSEE162 pKa = 4.06EE163 pKa = 3.72PAYY166 pKa = 10.52GSLWHH171 pKa = 5.94EE172 pKa = 4.57TSHH175 pKa = 7.26EE176 pKa = 4.07MMEE179 pKa = 4.21
MM1 pKa = 7.57ASSLVLEE8 pKa = 4.63SRR10 pKa = 11.84GIANNVKK17 pKa = 9.98LHH19 pKa = 6.67EE20 pKa = 4.9DD21 pKa = 3.51FEE23 pKa = 6.56GIDD26 pKa = 3.76VEE28 pKa = 4.34VLEE31 pKa = 4.95INSSILKK38 pKa = 10.42SLLEE42 pKa = 4.39DD43 pKa = 4.11AQGDD47 pKa = 4.07DD48 pKa = 4.31QDD50 pKa = 4.28YY51 pKa = 11.17DD52 pKa = 3.44QLNDD56 pKa = 3.5VIRR59 pKa = 11.84SLEE62 pKa = 3.93AEE64 pKa = 4.23IDD66 pKa = 3.75PSSLAGHH73 pKa = 7.37DD74 pKa = 4.25LGTHH78 pKa = 5.31TKK80 pKa = 10.33VINDD84 pKa = 4.04LVAGDD89 pKa = 4.25EE90 pKa = 4.06NSTINSRR97 pKa = 11.84GRR99 pKa = 11.84DD100 pKa = 3.54GSRR103 pKa = 11.84DD104 pKa = 3.0WSEE107 pKa = 3.91YY108 pKa = 10.51SSDD111 pKa = 3.54DD112 pKa = 4.09FSWIDD117 pKa = 3.42MDD119 pKa = 4.61TVPSSPGEE127 pKa = 4.0GMDD130 pKa = 2.81WYY132 pKa = 10.36MEE134 pKa = 4.14EE135 pKa = 5.55FGDD138 pKa = 5.23EE139 pKa = 4.49MNCLTQFGCIGDD151 pKa = 3.96YY152 pKa = 10.05YY153 pKa = 10.74QYY155 pKa = 9.6PQVYY159 pKa = 9.7SSEE162 pKa = 4.06EE163 pKa = 3.72PAYY166 pKa = 10.52GSLWHH171 pKa = 5.94EE172 pKa = 4.57TSHH175 pKa = 7.26EE176 pKa = 4.07MMEE179 pKa = 4.21
Molecular weight: 20.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2I0HYL5|A0A2I0HYL5_PUNGR Uncharacterized protein OS=Punica granatum OX=22663 GN=CRG98_042983 PE=4 SV=1
MM1 pKa = 8.03DD2 pKa = 4.85FASTLPFSTDD12 pKa = 3.33FGLVTLRR19 pKa = 11.84LSTGFGLVTLRR30 pKa = 11.84LSTGFGLVTLRR41 pKa = 11.84LSTGFGLVTLRR52 pKa = 11.84LSTGFGLVTLRR63 pKa = 11.84LSTGFGLVTLRR74 pKa = 11.84LSTGFGLVTLRR85 pKa = 11.84LSTGFGLVTLRR96 pKa = 11.84LSTGFGLVTLRR107 pKa = 11.84LSTGFGLVTLRR118 pKa = 11.84LSTGFGLVTLRR129 pKa = 11.84LSTGFGLVTLRR140 pKa = 11.84LSTGFGLVTLRR151 pKa = 11.84LSTGFGLVTLRR162 pKa = 11.84LSTGFGLVTLRR173 pKa = 11.84LSTGFGLVTLRR184 pKa = 11.84LSTGFGLVTLRR195 pKa = 11.84LSTGFGLVTLRR206 pKa = 11.84LSTGFGLVTLRR217 pKa = 11.84LSTGFGLVTLRR228 pKa = 11.84LSTGFGLVTLRR239 pKa = 11.84LSTGFGLVTLRR250 pKa = 11.84LSTGFGLVTLRR261 pKa = 11.84LSTGFGLVTLRR272 pKa = 11.84LSTGFGLVTLRR283 pKa = 11.84LSTGFGLVTLRR294 pKa = 11.84LSTGFGLVTLRR305 pKa = 11.84LSTGFGLVTLRR316 pKa = 11.84LSTGFGLVTLRR327 pKa = 11.84LSTGFGLVTLRR338 pKa = 11.84LSTGFGLVTLRR349 pKa = 11.84LSTGFGLVTLRR360 pKa = 11.84LSTGFGLVTLRR371 pKa = 11.84LSTGFGLVTLRR382 pKa = 11.84LSTGFGLVTLRR393 pKa = 11.84LSTGFGLVTLRR404 pKa = 11.84LSTGFGLVTLRR415 pKa = 11.84LSTGFGLVTLRR426 pKa = 11.84LSTGFGLVTLRR437 pKa = 11.84LSTGFGLVTLRR448 pKa = 11.84LSTGFGLVTLRR459 pKa = 11.84LSTGFGLVTLRR470 pKa = 11.84LSTGFGLVTLRR481 pKa = 11.84LSTGFGLVTLRR492 pKa = 11.84LSTGFGLVTLRR503 pKa = 11.84LSTGFGLVTLRR514 pKa = 11.84LSTGFGLVTLRR525 pKa = 11.84LSTGFGLVTLRR536 pKa = 11.84LSTGFGLVTLRR547 pKa = 11.84LSTGFGLVTLRR558 pKa = 11.84LSTGFGLVTLRR569 pKa = 11.84LSTGFGLVTLRR580 pKa = 11.84LSTGFGLVTLRR591 pKa = 11.84LSTGFGLVTLRR602 pKa = 11.84LSTGFGLVTLRR613 pKa = 11.84LSTGFGLVTLRR624 pKa = 11.84LSTGFGLVTLRR635 pKa = 11.84LSTGFGLVTLRR646 pKa = 11.84LSTGFGLVTLRR657 pKa = 11.84LSTGFGLVTLRR668 pKa = 11.84LSTGFGLVTLRR679 pKa = 11.84LSTGFGLVTLRR690 pKa = 11.84LSTGFGLVTLRR701 pKa = 11.84LSTGFGLVTLRR712 pKa = 11.84LSTGFGLVTLRR723 pKa = 11.84LSTGFGLVTLRR734 pKa = 11.84LSTGFGLVTLRR745 pKa = 11.84LSTGFGLVTLRR756 pKa = 11.84LSTGFGLVTLRR767 pKa = 11.84LSTGFGLVTLPFSTDD782 pKa = 3.4FGLVTLRR789 pKa = 11.84LSTGFGLVTLLLSEE803 pKa = 4.84LGVHH807 pKa = 6.68GLRR810 pKa = 11.84IHH812 pKa = 5.82FTVFHH817 pKa = 6.73GLRR820 pKa = 11.84PSNFTVFHH828 pKa = 6.6GLRR831 pKa = 11.84PGNITAFRR839 pKa = 11.84GLRR842 pKa = 11.84LSKK845 pKa = 10.42FIAFRR850 pKa = 11.84TSRR853 pKa = 11.84PWTSRR858 pKa = 11.84PLYY861 pKa = 9.64RR862 pKa = 11.84FPRR865 pKa = 11.84ASAA868 pKa = 3.27
MM1 pKa = 8.03DD2 pKa = 4.85FASTLPFSTDD12 pKa = 3.33FGLVTLRR19 pKa = 11.84LSTGFGLVTLRR30 pKa = 11.84LSTGFGLVTLRR41 pKa = 11.84LSTGFGLVTLRR52 pKa = 11.84LSTGFGLVTLRR63 pKa = 11.84LSTGFGLVTLRR74 pKa = 11.84LSTGFGLVTLRR85 pKa = 11.84LSTGFGLVTLRR96 pKa = 11.84LSTGFGLVTLRR107 pKa = 11.84LSTGFGLVTLRR118 pKa = 11.84LSTGFGLVTLRR129 pKa = 11.84LSTGFGLVTLRR140 pKa = 11.84LSTGFGLVTLRR151 pKa = 11.84LSTGFGLVTLRR162 pKa = 11.84LSTGFGLVTLRR173 pKa = 11.84LSTGFGLVTLRR184 pKa = 11.84LSTGFGLVTLRR195 pKa = 11.84LSTGFGLVTLRR206 pKa = 11.84LSTGFGLVTLRR217 pKa = 11.84LSTGFGLVTLRR228 pKa = 11.84LSTGFGLVTLRR239 pKa = 11.84LSTGFGLVTLRR250 pKa = 11.84LSTGFGLVTLRR261 pKa = 11.84LSTGFGLVTLRR272 pKa = 11.84LSTGFGLVTLRR283 pKa = 11.84LSTGFGLVTLRR294 pKa = 11.84LSTGFGLVTLRR305 pKa = 11.84LSTGFGLVTLRR316 pKa = 11.84LSTGFGLVTLRR327 pKa = 11.84LSTGFGLVTLRR338 pKa = 11.84LSTGFGLVTLRR349 pKa = 11.84LSTGFGLVTLRR360 pKa = 11.84LSTGFGLVTLRR371 pKa = 11.84LSTGFGLVTLRR382 pKa = 11.84LSTGFGLVTLRR393 pKa = 11.84LSTGFGLVTLRR404 pKa = 11.84LSTGFGLVTLRR415 pKa = 11.84LSTGFGLVTLRR426 pKa = 11.84LSTGFGLVTLRR437 pKa = 11.84LSTGFGLVTLRR448 pKa = 11.84LSTGFGLVTLRR459 pKa = 11.84LSTGFGLVTLRR470 pKa = 11.84LSTGFGLVTLRR481 pKa = 11.84LSTGFGLVTLRR492 pKa = 11.84LSTGFGLVTLRR503 pKa = 11.84LSTGFGLVTLRR514 pKa = 11.84LSTGFGLVTLRR525 pKa = 11.84LSTGFGLVTLRR536 pKa = 11.84LSTGFGLVTLRR547 pKa = 11.84LSTGFGLVTLRR558 pKa = 11.84LSTGFGLVTLRR569 pKa = 11.84LSTGFGLVTLRR580 pKa = 11.84LSTGFGLVTLRR591 pKa = 11.84LSTGFGLVTLRR602 pKa = 11.84LSTGFGLVTLRR613 pKa = 11.84LSTGFGLVTLRR624 pKa = 11.84LSTGFGLVTLRR635 pKa = 11.84LSTGFGLVTLRR646 pKa = 11.84LSTGFGLVTLRR657 pKa = 11.84LSTGFGLVTLRR668 pKa = 11.84LSTGFGLVTLRR679 pKa = 11.84LSTGFGLVTLRR690 pKa = 11.84LSTGFGLVTLRR701 pKa = 11.84LSTGFGLVTLRR712 pKa = 11.84LSTGFGLVTLRR723 pKa = 11.84LSTGFGLVTLRR734 pKa = 11.84LSTGFGLVTLRR745 pKa = 11.84LSTGFGLVTLRR756 pKa = 11.84LSTGFGLVTLRR767 pKa = 11.84LSTGFGLVTLPFSTDD782 pKa = 3.4FGLVTLRR789 pKa = 11.84LSTGFGLVTLLLSEE803 pKa = 4.84LGVHH807 pKa = 6.68GLRR810 pKa = 11.84IHH812 pKa = 5.82FTVFHH817 pKa = 6.73GLRR820 pKa = 11.84PSNFTVFHH828 pKa = 6.6GLRR831 pKa = 11.84PGNITAFRR839 pKa = 11.84GLRR842 pKa = 11.84LSKK845 pKa = 10.42FIAFRR850 pKa = 11.84TSRR853 pKa = 11.84PWTSRR858 pKa = 11.84PLYY861 pKa = 9.64RR862 pKa = 11.84FPRR865 pKa = 11.84ASAA868 pKa = 3.27
Molecular weight: 91.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14680046 |
11 |
11234 |
291.1 |
32.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.176 ± 0.022 | 2.043 ± 0.015 |
4.916 ± 0.018 | 6.147 ± 0.023 |
4.216 ± 0.02 | 7.248 ± 0.031 |
2.463 ± 0.01 | 4.684 ± 0.017 |
5.168 ± 0.02 | 9.854 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.305 ± 0.009 | 3.592 ± 0.014 |
5.962 ± 0.021 | 3.345 ± 0.01 |
6.666 ± 0.022 | 8.957 ± 0.024 |
4.994 ± 0.013 | 6.35 ± 0.013 |
1.33 ± 0.007 | 2.578 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
