
Roseivirga misakiensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Roseivirgaceae; Roseivirga
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
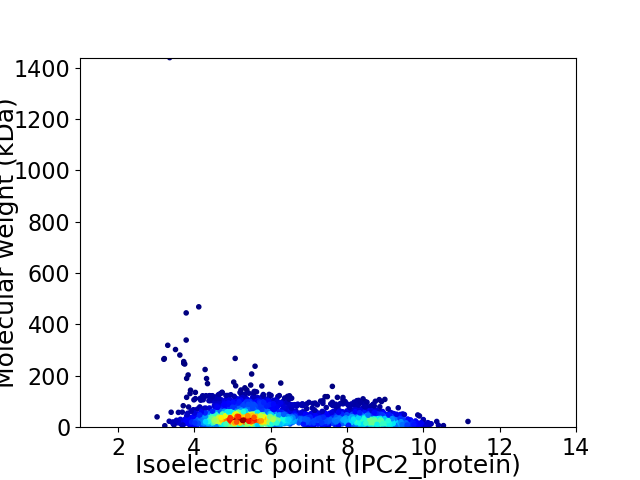
Virtual 2D-PAGE plot for 3764 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E5T5T3|A0A1E5T5T3_9BACT DnaB domain-containing protein OS=Roseivirga misakiensis OX=1563681 GN=BFP71_03395 PE=4 SV=1
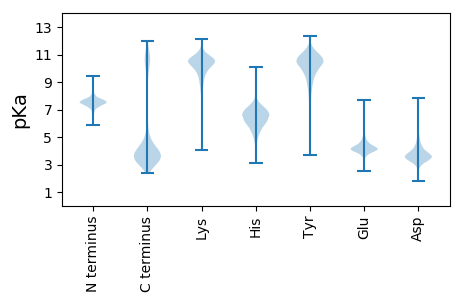
MM1 pKa = 7.75KK2 pKa = 10.49KK3 pKa = 10.06FLKK6 pKa = 10.13KK7 pKa = 10.92GMMGALMLSALVFTACDD24 pKa = 3.56NEE26 pKa = 4.62DD27 pKa = 4.75DD28 pKa = 4.99NGTDD32 pKa = 3.56PAGQDD37 pKa = 3.11VTVVLNEE44 pKa = 3.66VNYY47 pKa = 11.03ASGDD51 pKa = 3.42WIEE54 pKa = 4.21IFNNGSAPADD64 pKa = 3.54LSNYY68 pKa = 7.1WLCLGPGSYY77 pKa = 10.47VEE79 pKa = 5.07LSAITPQTGSVTNLLSGEE97 pKa = 4.27YY98 pKa = 10.19LVLPYY103 pKa = 10.42TMPDD107 pKa = 3.05TDD109 pKa = 4.48GGLGLYY115 pKa = 10.23SSNEE119 pKa = 4.04FTNSAAIVDD128 pKa = 4.57FVQWGSGGSARR139 pKa = 11.84EE140 pKa = 4.01DD141 pKa = 3.47VAVSAGLWTAGEE153 pKa = 4.26YY154 pKa = 10.87VPTVSSEE161 pKa = 4.05DD162 pKa = 3.26NSIIFDD168 pKa = 3.97GEE170 pKa = 4.35GNGAANWAEE179 pKa = 4.32TAQPTPGNEE188 pKa = 3.73NVLIVPEE195 pKa = 4.17PARR198 pKa = 11.84SIVISEE204 pKa = 3.97VQYY207 pKa = 11.2GNRR210 pKa = 11.84NLVEE214 pKa = 4.48LYY216 pKa = 11.1NNGEE220 pKa = 4.09VTIDD224 pKa = 3.71LSSYY228 pKa = 8.14WLCLGPGAYY237 pKa = 9.72AQIGNLTPEE246 pKa = 4.41FGSIQVEE253 pKa = 3.9AGDD256 pKa = 4.19FVVLPFNMPDD266 pKa = 3.54DD267 pKa = 4.35EE268 pKa = 6.77GGLGLYY274 pKa = 10.65SMNQFTSPDD283 pKa = 3.63AIMDD287 pKa = 4.0FVQWGATGSARR298 pKa = 11.84EE299 pKa = 4.16NVAVTAGVWTAGNFVPTVRR318 pKa = 11.84LDD320 pKa = 3.5SYY322 pKa = 11.13SIEE325 pKa = 4.1VNMEE329 pKa = 3.89SDD331 pKa = 3.77GSLASDD337 pKa = 3.05WSEE340 pKa = 3.57EE341 pKa = 4.15VNPSLGAPNNEE352 pKa = 3.81AVEE355 pKa = 4.42TTTFNVTISNMTNYY369 pKa = 10.48LNVHH373 pKa = 6.18TFTTPVGANSAGPLGVNGAQYY394 pKa = 10.53QIEE397 pKa = 4.53FKK399 pKa = 10.84AVAGTKK405 pKa = 7.43FTPVTMMGNSNDD417 pKa = 3.21WFLAPTDD424 pKa = 3.9LAGIDD429 pKa = 3.99LFPNGTALNSVDD441 pKa = 4.66IADD444 pKa = 3.87QLSLYY449 pKa = 10.87DD450 pKa = 5.26LGTEE454 pKa = 5.21ADD456 pKa = 3.61NDD458 pKa = 4.17PNNFPPAGTNVGPADD473 pKa = 3.31SDD475 pKa = 3.53ATVRR479 pKa = 11.84LVSGRR484 pKa = 11.84GTGADD489 pKa = 3.62YY490 pKa = 7.6MTAILTYY497 pKa = 10.58AAGAQNEE504 pKa = 4.53AGTFTLTITATNAPDD519 pKa = 3.64ANQAASQNNGFIITPGMVVLHH540 pKa = 6.73ALPEE544 pKa = 4.14PLFTLGSEE552 pKa = 4.05DD553 pKa = 3.25RR554 pKa = 11.84AVGLEE559 pKa = 4.21RR560 pKa = 11.84IAEE563 pKa = 4.26DD564 pKa = 3.67GMPSEE569 pKa = 4.81LYY571 pKa = 10.58DD572 pKa = 3.36WFKK575 pKa = 10.77EE576 pKa = 4.36TGSNGAPLRR585 pKa = 11.84LSSSLSVFSPGLVYY599 pKa = 10.91AFNTTTDD606 pKa = 3.68PLFTQGDD613 pKa = 3.76AARR616 pKa = 11.84AGSGIEE622 pKa = 3.96EE623 pKa = 4.23LAEE626 pKa = 4.77DD627 pKa = 4.72GNNQIAVEE635 pKa = 4.29YY636 pKa = 8.68ITNLGLPVAASNEE649 pKa = 4.35TVNIAPGEE657 pKa = 4.05DD658 pKa = 3.05LTFSIEE664 pKa = 4.02VPAGQNYY671 pKa = 10.13KK672 pKa = 10.76LGIGTMLVQTNDD684 pKa = 2.32WFVAYY689 pKa = 10.17NNSGVALWDD698 pKa = 3.72ANGAPTSGTSNSQRR712 pKa = 11.84TYY714 pKa = 10.83LYY716 pKa = 10.86DD717 pKa = 4.41AGTEE721 pKa = 3.86DD722 pKa = 4.8DD723 pKa = 3.9EE724 pKa = 5.46AVGFGANQAPRR735 pKa = 11.84QSGPNQGSADD745 pKa = 3.7GNTDD749 pKa = 2.43IRR751 pKa = 11.84RR752 pKa = 11.84VGEE755 pKa = 4.07LEE757 pKa = 3.94DD758 pKa = 3.54VQFGKK763 pKa = 11.13GLITNGPGTTYY774 pKa = 10.78LRR776 pKa = 11.84DD777 pKa = 3.29PRR779 pKa = 11.84GGYY782 pKa = 10.18NIIRR786 pKa = 11.84VDD788 pKa = 3.35IQPNN792 pKa = 3.18
MM1 pKa = 7.75KK2 pKa = 10.49KK3 pKa = 10.06FLKK6 pKa = 10.13KK7 pKa = 10.92GMMGALMLSALVFTACDD24 pKa = 3.56NEE26 pKa = 4.62DD27 pKa = 4.75DD28 pKa = 4.99NGTDD32 pKa = 3.56PAGQDD37 pKa = 3.11VTVVLNEE44 pKa = 3.66VNYY47 pKa = 11.03ASGDD51 pKa = 3.42WIEE54 pKa = 4.21IFNNGSAPADD64 pKa = 3.54LSNYY68 pKa = 7.1WLCLGPGSYY77 pKa = 10.47VEE79 pKa = 5.07LSAITPQTGSVTNLLSGEE97 pKa = 4.27YY98 pKa = 10.19LVLPYY103 pKa = 10.42TMPDD107 pKa = 3.05TDD109 pKa = 4.48GGLGLYY115 pKa = 10.23SSNEE119 pKa = 4.04FTNSAAIVDD128 pKa = 4.57FVQWGSGGSARR139 pKa = 11.84EE140 pKa = 4.01DD141 pKa = 3.47VAVSAGLWTAGEE153 pKa = 4.26YY154 pKa = 10.87VPTVSSEE161 pKa = 4.05DD162 pKa = 3.26NSIIFDD168 pKa = 3.97GEE170 pKa = 4.35GNGAANWAEE179 pKa = 4.32TAQPTPGNEE188 pKa = 3.73NVLIVPEE195 pKa = 4.17PARR198 pKa = 11.84SIVISEE204 pKa = 3.97VQYY207 pKa = 11.2GNRR210 pKa = 11.84NLVEE214 pKa = 4.48LYY216 pKa = 11.1NNGEE220 pKa = 4.09VTIDD224 pKa = 3.71LSSYY228 pKa = 8.14WLCLGPGAYY237 pKa = 9.72AQIGNLTPEE246 pKa = 4.41FGSIQVEE253 pKa = 3.9AGDD256 pKa = 4.19FVVLPFNMPDD266 pKa = 3.54DD267 pKa = 4.35EE268 pKa = 6.77GGLGLYY274 pKa = 10.65SMNQFTSPDD283 pKa = 3.63AIMDD287 pKa = 4.0FVQWGATGSARR298 pKa = 11.84EE299 pKa = 4.16NVAVTAGVWTAGNFVPTVRR318 pKa = 11.84LDD320 pKa = 3.5SYY322 pKa = 11.13SIEE325 pKa = 4.1VNMEE329 pKa = 3.89SDD331 pKa = 3.77GSLASDD337 pKa = 3.05WSEE340 pKa = 3.57EE341 pKa = 4.15VNPSLGAPNNEE352 pKa = 3.81AVEE355 pKa = 4.42TTTFNVTISNMTNYY369 pKa = 10.48LNVHH373 pKa = 6.18TFTTPVGANSAGPLGVNGAQYY394 pKa = 10.53QIEE397 pKa = 4.53FKK399 pKa = 10.84AVAGTKK405 pKa = 7.43FTPVTMMGNSNDD417 pKa = 3.21WFLAPTDD424 pKa = 3.9LAGIDD429 pKa = 3.99LFPNGTALNSVDD441 pKa = 4.66IADD444 pKa = 3.87QLSLYY449 pKa = 10.87DD450 pKa = 5.26LGTEE454 pKa = 5.21ADD456 pKa = 3.61NDD458 pKa = 4.17PNNFPPAGTNVGPADD473 pKa = 3.31SDD475 pKa = 3.53ATVRR479 pKa = 11.84LVSGRR484 pKa = 11.84GTGADD489 pKa = 3.62YY490 pKa = 7.6MTAILTYY497 pKa = 10.58AAGAQNEE504 pKa = 4.53AGTFTLTITATNAPDD519 pKa = 3.64ANQAASQNNGFIITPGMVVLHH540 pKa = 6.73ALPEE544 pKa = 4.14PLFTLGSEE552 pKa = 4.05DD553 pKa = 3.25RR554 pKa = 11.84AVGLEE559 pKa = 4.21RR560 pKa = 11.84IAEE563 pKa = 4.26DD564 pKa = 3.67GMPSEE569 pKa = 4.81LYY571 pKa = 10.58DD572 pKa = 3.36WFKK575 pKa = 10.77EE576 pKa = 4.36TGSNGAPLRR585 pKa = 11.84LSSSLSVFSPGLVYY599 pKa = 10.91AFNTTTDD606 pKa = 3.68PLFTQGDD613 pKa = 3.76AARR616 pKa = 11.84AGSGIEE622 pKa = 3.96EE623 pKa = 4.23LAEE626 pKa = 4.77DD627 pKa = 4.72GNNQIAVEE635 pKa = 4.29YY636 pKa = 8.68ITNLGLPVAASNEE649 pKa = 4.35TVNIAPGEE657 pKa = 4.05DD658 pKa = 3.05LTFSIEE664 pKa = 4.02VPAGQNYY671 pKa = 10.13KK672 pKa = 10.76LGIGTMLVQTNDD684 pKa = 2.32WFVAYY689 pKa = 10.17NNSGVALWDD698 pKa = 3.72ANGAPTSGTSNSQRR712 pKa = 11.84TYY714 pKa = 10.83LYY716 pKa = 10.86DD717 pKa = 4.41AGTEE721 pKa = 3.86DD722 pKa = 4.8DD723 pKa = 3.9EE724 pKa = 5.46AVGFGANQAPRR735 pKa = 11.84QSGPNQGSADD745 pKa = 3.7GNTDD749 pKa = 2.43IRR751 pKa = 11.84RR752 pKa = 11.84VGEE755 pKa = 4.07LEE757 pKa = 3.94DD758 pKa = 3.54VQFGKK763 pKa = 11.13GLITNGPGTTYY774 pKa = 10.78LRR776 pKa = 11.84DD777 pKa = 3.29PRR779 pKa = 11.84GGYY782 pKa = 10.18NIIRR786 pKa = 11.84VDD788 pKa = 3.35IQPNN792 pKa = 3.18
Molecular weight: 83.49 kDa
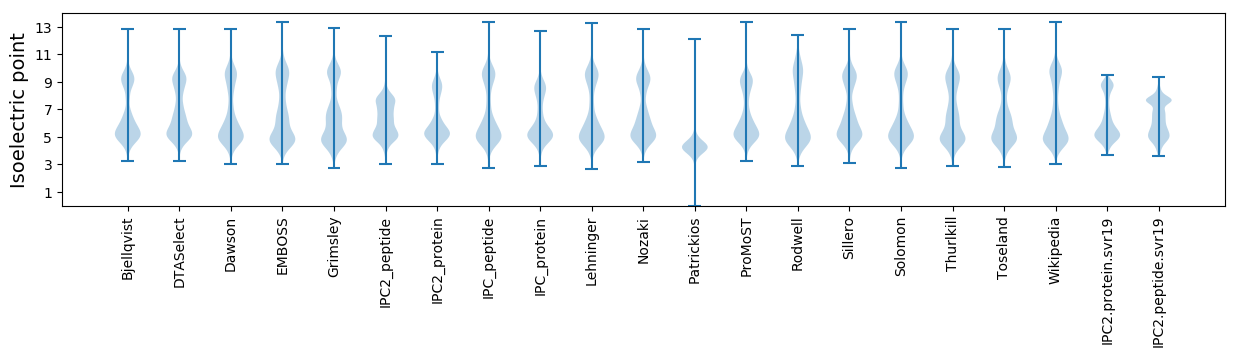
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E5T2V2|A0A1E5T2V2_9BACT Adenylosuccinate synthetase OS=Roseivirga misakiensis OX=1563681 GN=purA PE=3 SV=1
MM1 pKa = 7.36NGVTPTRR8 pKa = 11.84SQRR11 pKa = 11.84LSSTSGNSRR20 pKa = 11.84SAVTRR25 pKa = 11.84SAIASRR31 pKa = 11.84SQRR34 pKa = 11.84SSGRR38 pKa = 11.84VRR40 pKa = 11.84SSSSSGVSSSRR51 pKa = 11.84SSTRR55 pKa = 11.84NSTSSVASTRR65 pKa = 11.84STRR68 pKa = 11.84SRR70 pKa = 11.84RR71 pKa = 11.84TSTDD75 pKa = 2.17AYY77 pKa = 10.81RR78 pKa = 11.84FDD80 pKa = 4.15DD81 pKa = 3.46SSRR84 pKa = 11.84SRR86 pKa = 11.84RR87 pKa = 11.84SSSVRR92 pKa = 11.84ANATSGRR99 pKa = 11.84SAVSSRR105 pKa = 11.84TSRR108 pKa = 11.84SSSSRR113 pKa = 11.84SSVSRR118 pKa = 11.84SSSSRR123 pKa = 11.84SSSSRR128 pKa = 11.84PTFSRR133 pKa = 11.84SSSSRR138 pKa = 11.84SSSVRR143 pKa = 11.84SSSSRR148 pKa = 11.84SSSSRR153 pKa = 11.84SSVSRR158 pKa = 11.84SSSRR162 pKa = 11.84SSSSSRR168 pKa = 11.84SSGVRR173 pKa = 11.84SSSSRR178 pKa = 11.84SSSRR182 pKa = 11.84SSGVSRR188 pKa = 11.84SSSSRR193 pKa = 11.84SSSGSSRR200 pKa = 11.84SSSSSRR206 pKa = 11.84SGGSRR211 pKa = 11.84SSGRR215 pKa = 11.84GNN217 pKa = 3.16
MM1 pKa = 7.36NGVTPTRR8 pKa = 11.84SQRR11 pKa = 11.84LSSTSGNSRR20 pKa = 11.84SAVTRR25 pKa = 11.84SAIASRR31 pKa = 11.84SQRR34 pKa = 11.84SSGRR38 pKa = 11.84VRR40 pKa = 11.84SSSSSGVSSSRR51 pKa = 11.84SSTRR55 pKa = 11.84NSTSSVASTRR65 pKa = 11.84STRR68 pKa = 11.84SRR70 pKa = 11.84RR71 pKa = 11.84TSTDD75 pKa = 2.17AYY77 pKa = 10.81RR78 pKa = 11.84FDD80 pKa = 4.15DD81 pKa = 3.46SSRR84 pKa = 11.84SRR86 pKa = 11.84RR87 pKa = 11.84SSSVRR92 pKa = 11.84ANATSGRR99 pKa = 11.84SAVSSRR105 pKa = 11.84TSRR108 pKa = 11.84SSSSRR113 pKa = 11.84SSVSRR118 pKa = 11.84SSSSRR123 pKa = 11.84SSSSRR128 pKa = 11.84PTFSRR133 pKa = 11.84SSSSRR138 pKa = 11.84SSSVRR143 pKa = 11.84SSSSRR148 pKa = 11.84SSSSRR153 pKa = 11.84SSVSRR158 pKa = 11.84SSSRR162 pKa = 11.84SSSSSRR168 pKa = 11.84SSGVRR173 pKa = 11.84SSSSRR178 pKa = 11.84SSSRR182 pKa = 11.84SSGVSRR188 pKa = 11.84SSSSRR193 pKa = 11.84SSSGSSRR200 pKa = 11.84SSSSSRR206 pKa = 11.84SGGSRR211 pKa = 11.84SSGRR215 pKa = 11.84GNN217 pKa = 3.16
Molecular weight: 22.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1367009 |
52 |
14036 |
363.2 |
40.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.856 ± 0.048 | 0.591 ± 0.013 |
5.909 ± 0.053 | 6.619 ± 0.052 |
5.211 ± 0.034 | 6.913 ± 0.036 |
1.654 ± 0.024 | 7.362 ± 0.04 |
6.607 ± 0.072 | 9.591 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.28 ± 0.029 | 5.467 ± 0.052 |
3.518 ± 0.022 | 3.507 ± 0.021 |
4.07 ± 0.04 | 6.85 ± 0.042 |
5.612 ± 0.101 | 6.509 ± 0.041 |
1.119 ± 0.016 | 3.755 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
