
Mycoplasma phage MAV1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified dsDNA phages
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
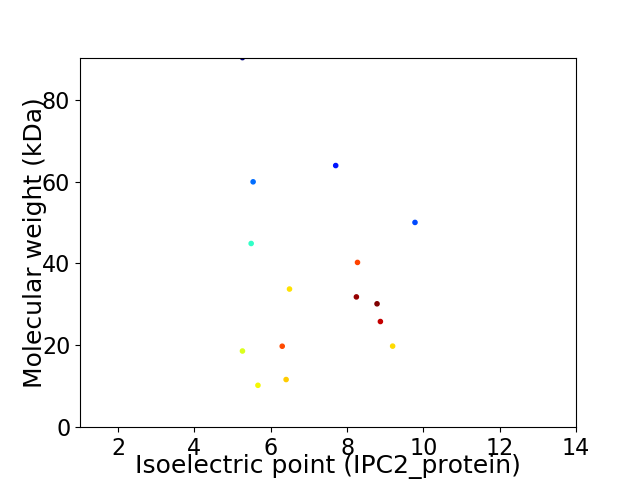
Virtual 2D-PAGE plot for 15 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|O80246|O80246_9VIRU HtpH OS=Mycoplasma phage MAV1 OX=75590 GN=htpH PE=4 SV=1
MM1 pKa = 7.47NDD3 pKa = 3.09KK4 pKa = 11.41NEE6 pKa = 4.21LLKK9 pKa = 11.19EE10 pKa = 4.0MGKK13 pKa = 7.97MASYY17 pKa = 10.13FSQLLARR24 pKa = 11.84SSKK27 pKa = 10.59SLQSEE32 pKa = 4.22LADD35 pKa = 3.94GADD38 pKa = 3.3VCRR41 pKa = 11.84LKK43 pKa = 10.31EE44 pKa = 4.25TIKK47 pKa = 10.98SLNEE51 pKa = 3.56DD52 pKa = 5.45ANWLWNKK59 pKa = 10.38LEE61 pKa = 4.62EE62 pKa = 3.97NWIMIRR68 pKa = 11.84LRR70 pKa = 11.84DD71 pKa = 3.69YY72 pKa = 11.36VDD74 pKa = 3.59YY75 pKa = 10.69EE76 pKa = 4.54GKK78 pKa = 8.16ITLYY82 pKa = 10.11KK83 pKa = 9.56YY84 pKa = 10.8KK85 pKa = 10.87VMNLTPNKK93 pKa = 9.69SLKK96 pKa = 9.83IHH98 pKa = 6.93LEE100 pKa = 4.01LNDD103 pKa = 4.94LIILEE108 pKa = 4.43PSSEE112 pKa = 3.51ISGYY116 pKa = 10.42IKK118 pKa = 10.58DD119 pKa = 3.48SARR122 pKa = 11.84NIIEE126 pKa = 3.87KK127 pKa = 10.38SGEE130 pKa = 4.05DD131 pKa = 3.54FSTFTTIDD139 pKa = 3.25SFNFEE144 pKa = 4.01TNEE147 pKa = 3.89PVFEE151 pKa = 4.32PLEE154 pKa = 4.18EE155 pKa = 4.06QDD157 pKa = 3.25QSFYY161 pKa = 11.1LYY163 pKa = 10.88SLNPKK168 pKa = 8.6ATINFKK174 pKa = 10.5RR175 pKa = 11.84DD176 pKa = 2.65NFYY179 pKa = 11.29YY180 pKa = 9.85IDD182 pKa = 4.01SNSLVEE188 pKa = 4.24YY189 pKa = 10.48QWTDD193 pKa = 3.5DD194 pKa = 4.26DD195 pKa = 4.74DD196 pKa = 4.5EE197 pKa = 6.57NYY199 pKa = 10.83LFTINFSNVKK209 pKa = 8.38QLKK212 pKa = 10.16NYY214 pKa = 8.5DD215 pKa = 3.43ASTIQYY221 pKa = 9.31KK222 pKa = 9.3WDD224 pKa = 3.63SEE226 pKa = 3.99ISEE229 pKa = 4.33EE230 pKa = 4.2STNTVEE236 pKa = 4.96LTFEE240 pKa = 4.48PSTLISDD247 pKa = 4.15LVFVGRR253 pKa = 11.84KK254 pKa = 8.84KK255 pKa = 11.14DD256 pKa = 3.4NDD258 pKa = 3.43KK259 pKa = 10.78TINNYY264 pKa = 9.43VSSARR269 pKa = 11.84LVNDD273 pKa = 3.44VYY275 pKa = 10.64FYY277 pKa = 10.48PHH279 pKa = 7.35LMNPIHH285 pKa = 5.5VQKK288 pKa = 10.95DD289 pKa = 3.58YY290 pKa = 11.19IQWNLPMLEE299 pKa = 3.34IVAYY303 pKa = 9.1VNRR306 pKa = 11.84LLKK309 pKa = 10.78DD310 pKa = 3.52AVDD313 pKa = 3.67VEE315 pKa = 4.68YY316 pKa = 10.51TSLRR320 pKa = 11.84TNILNIFSEE329 pKa = 4.32KK330 pKa = 10.57VINKK334 pKa = 7.25EE335 pKa = 3.83TLADD339 pKa = 4.67HH340 pKa = 6.06ITLFIDD346 pKa = 3.72TLFLCMDD353 pKa = 3.58VTKK356 pKa = 10.4KK357 pKa = 10.79VSSGYY362 pKa = 11.16AEE364 pKa = 4.34MGTAEE369 pKa = 4.23TNTFKK374 pKa = 10.57EE375 pKa = 4.61TVYY378 pKa = 10.86SGNNPKK384 pKa = 10.12EE385 pKa = 3.86FCDD388 pKa = 4.58YY389 pKa = 10.32KK390 pKa = 11.27LNWILQEE397 pKa = 4.06TEE399 pKa = 3.78KK400 pKa = 10.93FGNQSLIRR408 pKa = 11.84TFEE411 pKa = 4.74LIILMCSNQIASSYY425 pKa = 9.46CFKK428 pKa = 11.19GGLTTEE434 pKa = 4.24HH435 pKa = 6.59EE436 pKa = 4.83FYY438 pKa = 11.19LPFFIQSSYY447 pKa = 11.26KK448 pKa = 10.15NNKK451 pKa = 8.86NEE453 pKa = 3.76YY454 pKa = 9.17KK455 pKa = 10.01RR456 pKa = 11.84YY457 pKa = 9.6KK458 pKa = 10.37GSFVFKK464 pKa = 10.21SHH466 pKa = 6.06YY467 pKa = 9.73FALNEE472 pKa = 4.02TGQWQPLKK480 pKa = 10.81FDD482 pKa = 3.98INKK485 pKa = 9.75HH486 pKa = 4.2NARR489 pKa = 11.84FSKK492 pKa = 10.67NKK494 pKa = 9.06TLTFPLLPKK503 pKa = 10.43VLEE506 pKa = 4.21EE507 pKa = 4.2VNISNTPVDD516 pKa = 4.66FNTEE520 pKa = 3.2NSQNFKK526 pKa = 10.63YY527 pKa = 10.83LMWNVSEE534 pKa = 4.17QNHH537 pKa = 4.84VEE539 pKa = 4.21KK540 pKa = 10.92YY541 pKa = 10.87LKK543 pKa = 9.53TEE545 pKa = 4.2YY546 pKa = 10.53EE547 pKa = 4.3WIDD550 pKa = 3.27NGPTKK555 pKa = 10.31EE556 pKa = 4.41YY557 pKa = 10.37ILEE560 pKa = 4.19TEE562 pKa = 4.07PMEE565 pKa = 4.75IRR567 pKa = 11.84DD568 pKa = 3.63WNNAFDD574 pKa = 3.31IWLKK578 pKa = 9.2YY579 pKa = 10.38EE580 pKa = 3.91RR581 pKa = 11.84DD582 pKa = 3.45KK583 pKa = 10.61ITNYY587 pKa = 5.98EE588 pKa = 3.95TRR590 pKa = 11.84PILLRR595 pKa = 11.84AFKK598 pKa = 10.58DD599 pKa = 3.23NQNRR603 pKa = 11.84HH604 pKa = 6.14EE605 pKa = 4.34INKK608 pKa = 8.99EE609 pKa = 3.64QLLSILSKK617 pKa = 10.6PKK619 pKa = 10.64EE620 pKa = 3.96EE621 pKa = 4.67LKK623 pKa = 10.54YY624 pKa = 10.35LYY626 pKa = 10.53YY627 pKa = 10.59RR628 pKa = 11.84AFPTFFVSKK637 pKa = 9.78HH638 pKa = 4.58YY639 pKa = 10.57EE640 pKa = 3.86VLNVDD645 pKa = 4.04EE646 pKa = 4.77GAKK649 pKa = 8.82NTTAEE654 pKa = 4.23LFAYY658 pKa = 10.1DD659 pKa = 4.35PSTTNTPLFTYY670 pKa = 9.55FKK672 pKa = 10.22FRR674 pKa = 11.84SKK676 pKa = 10.93FKK678 pKa = 9.63ITSKK682 pKa = 8.76TQFRR686 pKa = 11.84DD687 pKa = 3.52DD688 pKa = 3.7YY689 pKa = 11.35QYY691 pKa = 7.82TTLIINPSEE700 pKa = 3.69FRR702 pKa = 11.84KK703 pKa = 10.45VINLPSDD710 pKa = 4.4PIKK713 pKa = 11.21VKK715 pKa = 11.0FEE717 pKa = 4.22LEE719 pKa = 3.49KK720 pKa = 11.29DD721 pKa = 4.35FILEE725 pKa = 4.35NITLKK730 pKa = 10.92SIFANQIGISLNGKK744 pKa = 6.08EE745 pKa = 4.19TKK747 pKa = 9.22TYY749 pKa = 9.68KK750 pKa = 10.57LNYY753 pKa = 9.16SSSISNTEE761 pKa = 3.62TKK763 pKa = 10.26IIII766 pKa = 4.07
MM1 pKa = 7.47NDD3 pKa = 3.09KK4 pKa = 11.41NEE6 pKa = 4.21LLKK9 pKa = 11.19EE10 pKa = 4.0MGKK13 pKa = 7.97MASYY17 pKa = 10.13FSQLLARR24 pKa = 11.84SSKK27 pKa = 10.59SLQSEE32 pKa = 4.22LADD35 pKa = 3.94GADD38 pKa = 3.3VCRR41 pKa = 11.84LKK43 pKa = 10.31EE44 pKa = 4.25TIKK47 pKa = 10.98SLNEE51 pKa = 3.56DD52 pKa = 5.45ANWLWNKK59 pKa = 10.38LEE61 pKa = 4.62EE62 pKa = 3.97NWIMIRR68 pKa = 11.84LRR70 pKa = 11.84DD71 pKa = 3.69YY72 pKa = 11.36VDD74 pKa = 3.59YY75 pKa = 10.69EE76 pKa = 4.54GKK78 pKa = 8.16ITLYY82 pKa = 10.11KK83 pKa = 9.56YY84 pKa = 10.8KK85 pKa = 10.87VMNLTPNKK93 pKa = 9.69SLKK96 pKa = 9.83IHH98 pKa = 6.93LEE100 pKa = 4.01LNDD103 pKa = 4.94LIILEE108 pKa = 4.43PSSEE112 pKa = 3.51ISGYY116 pKa = 10.42IKK118 pKa = 10.58DD119 pKa = 3.48SARR122 pKa = 11.84NIIEE126 pKa = 3.87KK127 pKa = 10.38SGEE130 pKa = 4.05DD131 pKa = 3.54FSTFTTIDD139 pKa = 3.25SFNFEE144 pKa = 4.01TNEE147 pKa = 3.89PVFEE151 pKa = 4.32PLEE154 pKa = 4.18EE155 pKa = 4.06QDD157 pKa = 3.25QSFYY161 pKa = 11.1LYY163 pKa = 10.88SLNPKK168 pKa = 8.6ATINFKK174 pKa = 10.5RR175 pKa = 11.84DD176 pKa = 2.65NFYY179 pKa = 11.29YY180 pKa = 9.85IDD182 pKa = 4.01SNSLVEE188 pKa = 4.24YY189 pKa = 10.48QWTDD193 pKa = 3.5DD194 pKa = 4.26DD195 pKa = 4.74DD196 pKa = 4.5EE197 pKa = 6.57NYY199 pKa = 10.83LFTINFSNVKK209 pKa = 8.38QLKK212 pKa = 10.16NYY214 pKa = 8.5DD215 pKa = 3.43ASTIQYY221 pKa = 9.31KK222 pKa = 9.3WDD224 pKa = 3.63SEE226 pKa = 3.99ISEE229 pKa = 4.33EE230 pKa = 4.2STNTVEE236 pKa = 4.96LTFEE240 pKa = 4.48PSTLISDD247 pKa = 4.15LVFVGRR253 pKa = 11.84KK254 pKa = 8.84KK255 pKa = 11.14DD256 pKa = 3.4NDD258 pKa = 3.43KK259 pKa = 10.78TINNYY264 pKa = 9.43VSSARR269 pKa = 11.84LVNDD273 pKa = 3.44VYY275 pKa = 10.64FYY277 pKa = 10.48PHH279 pKa = 7.35LMNPIHH285 pKa = 5.5VQKK288 pKa = 10.95DD289 pKa = 3.58YY290 pKa = 11.19IQWNLPMLEE299 pKa = 3.34IVAYY303 pKa = 9.1VNRR306 pKa = 11.84LLKK309 pKa = 10.78DD310 pKa = 3.52AVDD313 pKa = 3.67VEE315 pKa = 4.68YY316 pKa = 10.51TSLRR320 pKa = 11.84TNILNIFSEE329 pKa = 4.32KK330 pKa = 10.57VINKK334 pKa = 7.25EE335 pKa = 3.83TLADD339 pKa = 4.67HH340 pKa = 6.06ITLFIDD346 pKa = 3.72TLFLCMDD353 pKa = 3.58VTKK356 pKa = 10.4KK357 pKa = 10.79VSSGYY362 pKa = 11.16AEE364 pKa = 4.34MGTAEE369 pKa = 4.23TNTFKK374 pKa = 10.57EE375 pKa = 4.61TVYY378 pKa = 10.86SGNNPKK384 pKa = 10.12EE385 pKa = 3.86FCDD388 pKa = 4.58YY389 pKa = 10.32KK390 pKa = 11.27LNWILQEE397 pKa = 4.06TEE399 pKa = 3.78KK400 pKa = 10.93FGNQSLIRR408 pKa = 11.84TFEE411 pKa = 4.74LIILMCSNQIASSYY425 pKa = 9.46CFKK428 pKa = 11.19GGLTTEE434 pKa = 4.24HH435 pKa = 6.59EE436 pKa = 4.83FYY438 pKa = 11.19LPFFIQSSYY447 pKa = 11.26KK448 pKa = 10.15NNKK451 pKa = 8.86NEE453 pKa = 3.76YY454 pKa = 9.17KK455 pKa = 10.01RR456 pKa = 11.84YY457 pKa = 9.6KK458 pKa = 10.37GSFVFKK464 pKa = 10.21SHH466 pKa = 6.06YY467 pKa = 9.73FALNEE472 pKa = 4.02TGQWQPLKK480 pKa = 10.81FDD482 pKa = 3.98INKK485 pKa = 9.75HH486 pKa = 4.2NARR489 pKa = 11.84FSKK492 pKa = 10.67NKK494 pKa = 9.06TLTFPLLPKK503 pKa = 10.43VLEE506 pKa = 4.21EE507 pKa = 4.2VNISNTPVDD516 pKa = 4.66FNTEE520 pKa = 3.2NSQNFKK526 pKa = 10.63YY527 pKa = 10.83LMWNVSEE534 pKa = 4.17QNHH537 pKa = 4.84VEE539 pKa = 4.21KK540 pKa = 10.92YY541 pKa = 10.87LKK543 pKa = 9.53TEE545 pKa = 4.2YY546 pKa = 10.53EE547 pKa = 4.3WIDD550 pKa = 3.27NGPTKK555 pKa = 10.31EE556 pKa = 4.41YY557 pKa = 10.37ILEE560 pKa = 4.19TEE562 pKa = 4.07PMEE565 pKa = 4.75IRR567 pKa = 11.84DD568 pKa = 3.63WNNAFDD574 pKa = 3.31IWLKK578 pKa = 9.2YY579 pKa = 10.38EE580 pKa = 3.91RR581 pKa = 11.84DD582 pKa = 3.45KK583 pKa = 10.61ITNYY587 pKa = 5.98EE588 pKa = 3.95TRR590 pKa = 11.84PILLRR595 pKa = 11.84AFKK598 pKa = 10.58DD599 pKa = 3.23NQNRR603 pKa = 11.84HH604 pKa = 6.14EE605 pKa = 4.34INKK608 pKa = 8.99EE609 pKa = 3.64QLLSILSKK617 pKa = 10.6PKK619 pKa = 10.64EE620 pKa = 3.96EE621 pKa = 4.67LKK623 pKa = 10.54YY624 pKa = 10.35LYY626 pKa = 10.53YY627 pKa = 10.59RR628 pKa = 11.84AFPTFFVSKK637 pKa = 9.78HH638 pKa = 4.58YY639 pKa = 10.57EE640 pKa = 3.86VLNVDD645 pKa = 4.04EE646 pKa = 4.77GAKK649 pKa = 8.82NTTAEE654 pKa = 4.23LFAYY658 pKa = 10.1DD659 pKa = 4.35PSTTNTPLFTYY670 pKa = 9.55FKK672 pKa = 10.22FRR674 pKa = 11.84SKK676 pKa = 10.93FKK678 pKa = 9.63ITSKK682 pKa = 8.76TQFRR686 pKa = 11.84DD687 pKa = 3.52DD688 pKa = 3.7YY689 pKa = 11.35QYY691 pKa = 7.82TTLIINPSEE700 pKa = 3.69FRR702 pKa = 11.84KK703 pKa = 10.45VINLPSDD710 pKa = 4.4PIKK713 pKa = 11.21VKK715 pKa = 11.0FEE717 pKa = 4.22LEE719 pKa = 3.49KK720 pKa = 11.29DD721 pKa = 4.35FILEE725 pKa = 4.35NITLKK730 pKa = 10.92SIFANQIGISLNGKK744 pKa = 6.08EE745 pKa = 4.19TKK747 pKa = 9.22TYY749 pKa = 9.68KK750 pKa = 10.57LNYY753 pKa = 9.16SSSISNTEE761 pKa = 3.62TKK763 pKa = 10.26IIII766 pKa = 4.07
Molecular weight: 90.29 kDa
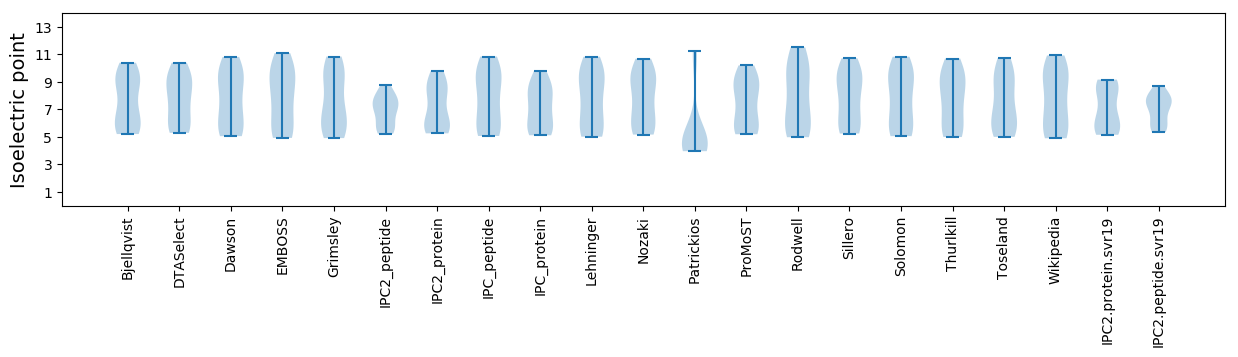
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|O80249|O80249_9VIRU HtpB OS=Mycoplasma phage MAV1 OX=75590 GN=htpB PE=4 SV=1
MM1 pKa = 7.61LSYY4 pKa = 10.83GKK6 pKa = 9.98NLISRR11 pKa = 11.84KK12 pKa = 9.76AIPLKK17 pKa = 10.18PKK19 pKa = 10.67SKK21 pKa = 10.41GKK23 pKa = 8.23PWWYY27 pKa = 9.34YY28 pKa = 10.38VVDD31 pKa = 3.96IVLDD35 pKa = 4.44LIIEE39 pKa = 4.33ALSLAVGVLSAGAASGITRR58 pKa = 11.84LGLGLIKK65 pKa = 10.52DD66 pKa = 3.67ITLSSVFDD74 pKa = 4.44RR75 pKa = 11.84KK76 pKa = 10.55INWKK80 pKa = 10.31DD81 pKa = 3.24VFLNAGLNLVFVGIGKK97 pKa = 9.37SLSKK101 pKa = 10.42FSKK104 pKa = 8.95ISKK107 pKa = 8.74SARR110 pKa = 11.84LIKK113 pKa = 10.52KK114 pKa = 8.52FAFYY118 pKa = 10.33AASPQKK124 pKa = 10.15FLNWIINKK132 pKa = 8.22ATYY135 pKa = 9.99GLKK138 pKa = 8.89KK139 pKa = 9.23TLINTVGQVAGKK151 pKa = 10.26KK152 pKa = 7.24ITKK155 pKa = 9.96FLRR158 pKa = 11.84TGLKK162 pKa = 10.22LISKK166 pKa = 7.06TAIITSAFLLAKK178 pKa = 10.35NKK180 pKa = 10.87AEE182 pKa = 3.92FAYY185 pKa = 10.09KK186 pKa = 10.29NIKK189 pKa = 8.51TFAIRR194 pKa = 11.84KK195 pKa = 7.5FKK197 pKa = 11.07NLLTKK202 pKa = 10.23SIRR205 pKa = 11.84RR206 pKa = 11.84QFTKK210 pKa = 10.84NFWGAIKK217 pKa = 10.33GKK219 pKa = 9.51KK220 pKa = 9.49LKK222 pKa = 10.76ASGYY226 pKa = 8.12NQILQKK232 pKa = 10.76HH233 pKa = 4.76NQTWIPFPSSKK244 pKa = 9.72WIEE247 pKa = 4.18GVKK250 pKa = 9.73IASDD254 pKa = 2.92NWIFNDD260 pKa = 3.52DD261 pKa = 3.65NTLISYY267 pKa = 8.59YY268 pKa = 11.03VFFKK272 pKa = 11.05SRR274 pKa = 11.84FAARR278 pKa = 11.84IRR280 pKa = 11.84TTVQKK285 pKa = 10.95DD286 pKa = 3.09PLIFIDD292 pKa = 4.84RR293 pKa = 11.84PIEE296 pKa = 3.94EE297 pKa = 4.66FNDD300 pKa = 3.78FLSSGSKK307 pKa = 10.3GKK309 pKa = 10.56FYY311 pKa = 10.83LDD313 pKa = 3.08EE314 pKa = 4.67LAWGWEE320 pKa = 3.78LGKK323 pKa = 10.54AIRR326 pKa = 11.84NQSKK330 pKa = 10.21KK331 pKa = 9.81PRR333 pKa = 11.84GTKK336 pKa = 10.07FYY338 pKa = 10.52INSLEE343 pKa = 4.77KK344 pKa = 9.42EE345 pKa = 4.11NKK347 pKa = 8.35YY348 pKa = 10.73NKK350 pKa = 9.95HH351 pKa = 5.56FLASLKK357 pKa = 10.82EE358 pKa = 4.01FDD360 pKa = 4.03TKK362 pKa = 11.54SNLAVEE368 pKa = 4.59EE369 pKa = 4.11FRR371 pKa = 11.84NQFKK375 pKa = 10.87VLSTTRR381 pKa = 11.84RR382 pKa = 11.84RR383 pKa = 11.84KK384 pKa = 9.64LGNKK388 pKa = 8.87IVVEE392 pKa = 4.84FKK394 pKa = 11.02SGDD397 pKa = 3.09TRR399 pKa = 11.84FYY401 pKa = 11.49GRR403 pKa = 11.84IPDD406 pKa = 3.51ATKK409 pKa = 10.46FNSKK413 pKa = 10.03FGNKK417 pKa = 9.16FQDD420 pKa = 3.43FRR422 pKa = 11.84KK423 pKa = 6.3TTKK426 pKa = 9.89NVKK429 pKa = 9.3LVKK432 pKa = 9.65TIKK435 pKa = 10.64KK436 pKa = 9.88LL437 pKa = 3.44
MM1 pKa = 7.61LSYY4 pKa = 10.83GKK6 pKa = 9.98NLISRR11 pKa = 11.84KK12 pKa = 9.76AIPLKK17 pKa = 10.18PKK19 pKa = 10.67SKK21 pKa = 10.41GKK23 pKa = 8.23PWWYY27 pKa = 9.34YY28 pKa = 10.38VVDD31 pKa = 3.96IVLDD35 pKa = 4.44LIIEE39 pKa = 4.33ALSLAVGVLSAGAASGITRR58 pKa = 11.84LGLGLIKK65 pKa = 10.52DD66 pKa = 3.67ITLSSVFDD74 pKa = 4.44RR75 pKa = 11.84KK76 pKa = 10.55INWKK80 pKa = 10.31DD81 pKa = 3.24VFLNAGLNLVFVGIGKK97 pKa = 9.37SLSKK101 pKa = 10.42FSKK104 pKa = 8.95ISKK107 pKa = 8.74SARR110 pKa = 11.84LIKK113 pKa = 10.52KK114 pKa = 8.52FAFYY118 pKa = 10.33AASPQKK124 pKa = 10.15FLNWIINKK132 pKa = 8.22ATYY135 pKa = 9.99GLKK138 pKa = 8.89KK139 pKa = 9.23TLINTVGQVAGKK151 pKa = 10.26KK152 pKa = 7.24ITKK155 pKa = 9.96FLRR158 pKa = 11.84TGLKK162 pKa = 10.22LISKK166 pKa = 7.06TAIITSAFLLAKK178 pKa = 10.35NKK180 pKa = 10.87AEE182 pKa = 3.92FAYY185 pKa = 10.09KK186 pKa = 10.29NIKK189 pKa = 8.51TFAIRR194 pKa = 11.84KK195 pKa = 7.5FKK197 pKa = 11.07NLLTKK202 pKa = 10.23SIRR205 pKa = 11.84RR206 pKa = 11.84QFTKK210 pKa = 10.84NFWGAIKK217 pKa = 10.33GKK219 pKa = 9.51KK220 pKa = 9.49LKK222 pKa = 10.76ASGYY226 pKa = 8.12NQILQKK232 pKa = 10.76HH233 pKa = 4.76NQTWIPFPSSKK244 pKa = 9.72WIEE247 pKa = 4.18GVKK250 pKa = 9.73IASDD254 pKa = 2.92NWIFNDD260 pKa = 3.52DD261 pKa = 3.65NTLISYY267 pKa = 8.59YY268 pKa = 11.03VFFKK272 pKa = 11.05SRR274 pKa = 11.84FAARR278 pKa = 11.84IRR280 pKa = 11.84TTVQKK285 pKa = 10.95DD286 pKa = 3.09PLIFIDD292 pKa = 4.84RR293 pKa = 11.84PIEE296 pKa = 3.94EE297 pKa = 4.66FNDD300 pKa = 3.78FLSSGSKK307 pKa = 10.3GKK309 pKa = 10.56FYY311 pKa = 10.83LDD313 pKa = 3.08EE314 pKa = 4.67LAWGWEE320 pKa = 3.78LGKK323 pKa = 10.54AIRR326 pKa = 11.84NQSKK330 pKa = 10.21KK331 pKa = 9.81PRR333 pKa = 11.84GTKK336 pKa = 10.07FYY338 pKa = 10.52INSLEE343 pKa = 4.77KK344 pKa = 9.42EE345 pKa = 4.11NKK347 pKa = 8.35YY348 pKa = 10.73NKK350 pKa = 9.95HH351 pKa = 5.56FLASLKK357 pKa = 10.82EE358 pKa = 4.01FDD360 pKa = 4.03TKK362 pKa = 11.54SNLAVEE368 pKa = 4.59EE369 pKa = 4.11FRR371 pKa = 11.84NQFKK375 pKa = 10.87VLSTTRR381 pKa = 11.84RR382 pKa = 11.84RR383 pKa = 11.84KK384 pKa = 9.64LGNKK388 pKa = 8.87IVVEE392 pKa = 4.84FKK394 pKa = 11.02SGDD397 pKa = 3.09TRR399 pKa = 11.84FYY401 pKa = 11.49GRR403 pKa = 11.84IPDD406 pKa = 3.51ATKK409 pKa = 10.46FNSKK413 pKa = 10.03FGNKK417 pKa = 9.16FQDD420 pKa = 3.43FRR422 pKa = 11.84KK423 pKa = 6.3TTKK426 pKa = 9.89NVKK429 pKa = 9.3LVKK432 pKa = 9.65TIKK435 pKa = 10.64KK436 pKa = 9.88LL437 pKa = 3.44
Molecular weight: 50.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4733 |
91 |
766 |
315.5 |
36.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.521 ± 0.457 | 0.739 ± 0.151 |
5.514 ± 0.42 | 7.057 ± 0.58 |
5.958 ± 0.299 | 3.528 ± 0.426 |
1.458 ± 0.143 | 8.684 ± 0.271 |
11.261 ± 0.567 | 9.719 ± 0.454 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.563 ± 0.204 | 8.494 ± 0.519 |
2.704 ± 0.218 | 3.444 ± 0.267 |
2.895 ± 0.286 | 6.529 ± 0.325 |
5.599 ± 0.523 | 4.606 ± 0.475 |
1.373 ± 0.182 | 4.352 ± 0.415 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
