
Roseiflexus sp. (strain RS-1)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Chloroflexia; Chloroflexales; Roseiflexineae; Roseiflexaceae; Roseiflexus; unclassified Roseiflexus
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
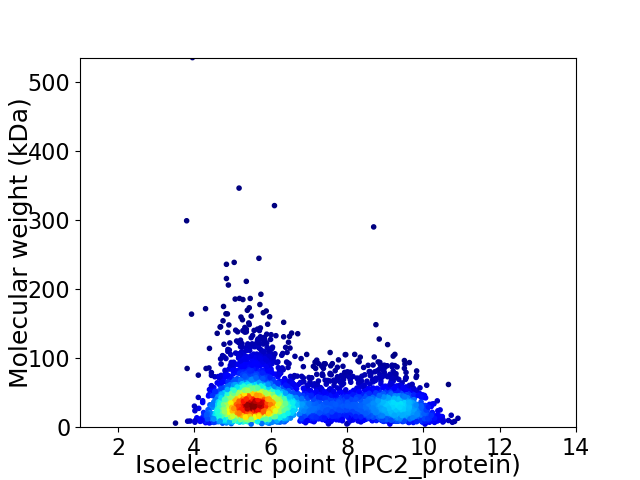
Virtual 2D-PAGE plot for 4494 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A5V179|A5V179_ROSS1 Uncharacterized protein OS=Roseiflexus sp. (strain RS-1) OX=357808 GN=RoseRS_4291 PE=4 SV=1
MM1 pKa = 7.23SASLKK6 pKa = 10.81SPATLFVLGVFLIMTAVLPAPRR28 pKa = 11.84PVSASSVLYY37 pKa = 9.97AAPTATGSADD47 pKa = 3.31CSSWANACTLQTALTNATSGNQIWVRR73 pKa = 11.84QGVHH77 pKa = 6.33RR78 pKa = 11.84PGSSQTDD85 pKa = 3.41TFTLKK90 pKa = 10.89NGVAVYY96 pKa = 10.52GGFAGTEE103 pKa = 3.78TSLAQRR109 pKa = 11.84DD110 pKa = 3.54WQTNVTVLSGDD121 pKa = 3.4INQDD125 pKa = 3.98DD126 pKa = 3.77ITDD129 pKa = 3.74ANGVVTSTANIAGVNAYY146 pKa = 9.87HH147 pKa = 6.17VVTGGGTNNTAVLDD161 pKa = 4.15GFTITAGNANSPSPPDD177 pKa = 3.6DD178 pKa = 4.49AGGGMYY184 pKa = 10.65NNNSSPTLRR193 pKa = 11.84NLIFSGNAANSGGGMANFNSSNPTLTNVTFSGNSADD229 pKa = 5.09LGGGMYY235 pKa = 10.48NEE237 pKa = 4.6NSSNPTLTNVTFSSNTAQSGGGMYY261 pKa = 10.54NFYY264 pKa = 10.94SSPTLTNVTFSGNAAQAGGGMFNSNSNPVLTNVTFSGNTASGMGSSGGGMFNYY317 pKa = 10.29DD318 pKa = 3.26NSSPTLTNVTFSSNTAQSGGGMYY341 pKa = 10.68NFFDD345 pKa = 4.69SSPTLTNVIFSGNAAQAGGGMFNEE369 pKa = 4.3NSSPILTNVIFSGNAVTVSGGGMANFNGSNPNLINVTFSGNTASSGGGMANFNGSNPTLTNVIIWNSVGGSIINSSSTPDD449 pKa = 3.17VTYY452 pKa = 11.41SNIQGGYY459 pKa = 10.09SGTTNINADD468 pKa = 4.07PFFVDD473 pKa = 5.16ADD475 pKa = 3.91GPDD478 pKa = 4.01NIVGTLDD485 pKa = 3.81DD486 pKa = 4.18NLRR489 pKa = 11.84LQSGSPSIDD498 pKa = 3.03TGNNAAVPTGVTTDD512 pKa = 3.95LEE514 pKa = 4.5GKK516 pKa = 9.52PRR518 pKa = 11.84IQNGTVDD525 pKa = 3.58MGAYY529 pKa = 9.6EE530 pKa = 4.45YY531 pKa = 11.04DD532 pKa = 3.52NTPPTVVSITRR543 pKa = 11.84ADD545 pKa = 3.8PNPTNAASVTFVVTFTEE562 pKa = 4.23AVTGVGVADD571 pKa = 4.9FALTLTGGVSGASVTGVSGSGAVYY595 pKa = 9.19TVTVSTGTGDD605 pKa = 3.09GTLRR609 pKa = 11.84LDD611 pKa = 3.76IPNTATITDD620 pKa = 4.17LAGNALSGLPFTGGQVYY637 pKa = 7.87TVDD640 pKa = 3.05KK641 pKa = 8.75TAPGVTMTSSAPNPTNSAPIPVTVTFSEE669 pKa = 4.62PVTGFTAGDD678 pKa = 3.37IAVSNGSVSNFAGGGASYY696 pKa = 9.93TFDD699 pKa = 3.82LTPAANGLVTATIAANVAVDD719 pKa = 3.33AAGNGNTAASFSRR732 pKa = 11.84TYY734 pKa = 10.38DD735 pKa = 3.07TTAPGVASITRR746 pKa = 11.84ADD748 pKa = 4.0PNPTSAASVNFIVTFTEE765 pKa = 3.94AVTGVGVADD774 pKa = 4.85FALTVTGISGASVTGVSGSGAIYY797 pKa = 9.46TVTVSTGTGDD807 pKa = 3.09GTLRR811 pKa = 11.84LDD813 pKa = 3.4IPTAATITDD822 pKa = 4.1LAGNALSGLPYY833 pKa = 10.31TSGEE837 pKa = 4.46TYY839 pKa = 9.9QKK841 pKa = 10.57GYY843 pKa = 10.83RR844 pKa = 11.84VFLPLIVRR852 pKa = 4.42
MM1 pKa = 7.23SASLKK6 pKa = 10.81SPATLFVLGVFLIMTAVLPAPRR28 pKa = 11.84PVSASSVLYY37 pKa = 9.97AAPTATGSADD47 pKa = 3.31CSSWANACTLQTALTNATSGNQIWVRR73 pKa = 11.84QGVHH77 pKa = 6.33RR78 pKa = 11.84PGSSQTDD85 pKa = 3.41TFTLKK90 pKa = 10.89NGVAVYY96 pKa = 10.52GGFAGTEE103 pKa = 3.78TSLAQRR109 pKa = 11.84DD110 pKa = 3.54WQTNVTVLSGDD121 pKa = 3.4INQDD125 pKa = 3.98DD126 pKa = 3.77ITDD129 pKa = 3.74ANGVVTSTANIAGVNAYY146 pKa = 9.87HH147 pKa = 6.17VVTGGGTNNTAVLDD161 pKa = 4.15GFTITAGNANSPSPPDD177 pKa = 3.6DD178 pKa = 4.49AGGGMYY184 pKa = 10.65NNNSSPTLRR193 pKa = 11.84NLIFSGNAANSGGGMANFNSSNPTLTNVTFSGNSADD229 pKa = 5.09LGGGMYY235 pKa = 10.48NEE237 pKa = 4.6NSSNPTLTNVTFSSNTAQSGGGMYY261 pKa = 10.54NFYY264 pKa = 10.94SSPTLTNVTFSGNAAQAGGGMFNSNSNPVLTNVTFSGNTASGMGSSGGGMFNYY317 pKa = 10.29DD318 pKa = 3.26NSSPTLTNVTFSSNTAQSGGGMYY341 pKa = 10.68NFFDD345 pKa = 4.69SSPTLTNVIFSGNAAQAGGGMFNEE369 pKa = 4.3NSSPILTNVIFSGNAVTVSGGGMANFNGSNPNLINVTFSGNTASSGGGMANFNGSNPTLTNVIIWNSVGGSIINSSSTPDD449 pKa = 3.17VTYY452 pKa = 11.41SNIQGGYY459 pKa = 10.09SGTTNINADD468 pKa = 4.07PFFVDD473 pKa = 5.16ADD475 pKa = 3.91GPDD478 pKa = 4.01NIVGTLDD485 pKa = 3.81DD486 pKa = 4.18NLRR489 pKa = 11.84LQSGSPSIDD498 pKa = 3.03TGNNAAVPTGVTTDD512 pKa = 3.95LEE514 pKa = 4.5GKK516 pKa = 9.52PRR518 pKa = 11.84IQNGTVDD525 pKa = 3.58MGAYY529 pKa = 9.6EE530 pKa = 4.45YY531 pKa = 11.04DD532 pKa = 3.52NTPPTVVSITRR543 pKa = 11.84ADD545 pKa = 3.8PNPTNAASVTFVVTFTEE562 pKa = 4.23AVTGVGVADD571 pKa = 4.9FALTLTGGVSGASVTGVSGSGAVYY595 pKa = 9.19TVTVSTGTGDD605 pKa = 3.09GTLRR609 pKa = 11.84LDD611 pKa = 3.76IPNTATITDD620 pKa = 4.17LAGNALSGLPFTGGQVYY637 pKa = 7.87TVDD640 pKa = 3.05KK641 pKa = 8.75TAPGVTMTSSAPNPTNSAPIPVTVTFSEE669 pKa = 4.62PVTGFTAGDD678 pKa = 3.37IAVSNGSVSNFAGGGASYY696 pKa = 9.93TFDD699 pKa = 3.82LTPAANGLVTATIAANVAVDD719 pKa = 3.33AAGNGNTAASFSRR732 pKa = 11.84TYY734 pKa = 10.38DD735 pKa = 3.07TTAPGVASITRR746 pKa = 11.84ADD748 pKa = 4.0PNPTSAASVNFIVTFTEE765 pKa = 3.94AVTGVGVADD774 pKa = 4.85FALTVTGISGASVTGVSGSGAIYY797 pKa = 9.46TVTVSTGTGDD807 pKa = 3.09GTLRR811 pKa = 11.84LDD813 pKa = 3.4IPTAATITDD822 pKa = 4.1LAGNALSGLPYY833 pKa = 10.31TSGEE837 pKa = 4.46TYY839 pKa = 9.9QKK841 pKa = 10.57GYY843 pKa = 10.83RR844 pKa = 11.84VFLPLIVRR852 pKa = 4.42
Molecular weight: 85.08 kDa
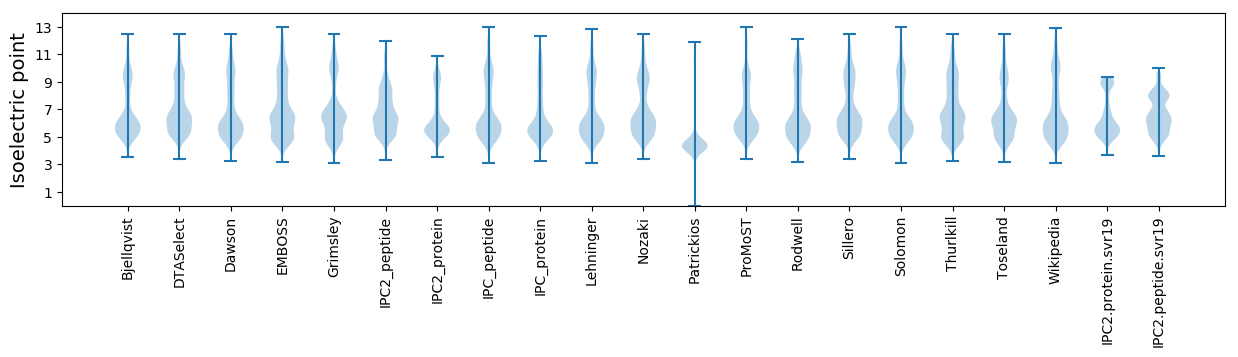
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A5UVQ2|A5UVQ2_ROSS1 Uncharacterized protein OS=Roseiflexus sp. (strain RS-1) OX=357808 GN=RoseRS_2326 PE=4 SV=1
MM1 pKa = 7.55CMSDD5 pKa = 4.12YY6 pKa = 10.67LTVAPPRR13 pKa = 11.84RR14 pKa = 11.84LRR16 pKa = 11.84LHH18 pKa = 6.82ASVQIAGMRR27 pKa = 11.84LPARR31 pKa = 11.84AFLMSVGILTAGGIAITLGADD52 pKa = 3.22IEE54 pKa = 4.37RR55 pKa = 11.84TIWASGCLIFLGLALLEE72 pKa = 4.1GRR74 pKa = 11.84VWGRR78 pKa = 11.84SSRR81 pKa = 11.84EE82 pKa = 3.37AAGIVWRR89 pKa = 11.84HH90 pKa = 5.25LKK92 pKa = 10.06RR93 pKa = 11.84PKK95 pKa = 9.97RR96 pKa = 11.84LRR98 pKa = 11.84LIQPHH103 pKa = 5.85MALPPEE109 pKa = 4.51EE110 pKa = 4.46ALSPVLKK117 pKa = 9.98PRR119 pKa = 11.84RR120 pKa = 11.84PHH122 pKa = 4.05WHH124 pKa = 5.17YY125 pKa = 11.22QEE127 pKa = 4.37TADD130 pKa = 4.63EE131 pKa = 4.43HH132 pKa = 6.99DD133 pKa = 3.48
MM1 pKa = 7.55CMSDD5 pKa = 4.12YY6 pKa = 10.67LTVAPPRR13 pKa = 11.84RR14 pKa = 11.84LRR16 pKa = 11.84LHH18 pKa = 6.82ASVQIAGMRR27 pKa = 11.84LPARR31 pKa = 11.84AFLMSVGILTAGGIAITLGADD52 pKa = 3.22IEE54 pKa = 4.37RR55 pKa = 11.84TIWASGCLIFLGLALLEE72 pKa = 4.1GRR74 pKa = 11.84VWGRR78 pKa = 11.84SSRR81 pKa = 11.84EE82 pKa = 3.37AAGIVWRR89 pKa = 11.84HH90 pKa = 5.25LKK92 pKa = 10.06RR93 pKa = 11.84PKK95 pKa = 9.97RR96 pKa = 11.84LRR98 pKa = 11.84LIQPHH103 pKa = 5.85MALPPEE109 pKa = 4.51EE110 pKa = 4.46ALSPVLKK117 pKa = 9.98PRR119 pKa = 11.84RR120 pKa = 11.84PHH122 pKa = 4.05WHH124 pKa = 5.17YY125 pKa = 11.22QEE127 pKa = 4.37TADD130 pKa = 4.63EE131 pKa = 4.43HH132 pKa = 6.99DD133 pKa = 3.48
Molecular weight: 14.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1633847 |
38 |
5166 |
363.6 |
39.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.859 ± 0.055 | 0.877 ± 0.012 |
5.445 ± 0.028 | 5.548 ± 0.041 |
3.328 ± 0.022 | 7.649 ± 0.037 |
2.18 ± 0.019 | 5.602 ± 0.026 |
1.852 ± 0.028 | 10.797 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.106 ± 0.014 | 2.494 ± 0.026 |
5.969 ± 0.032 | 3.651 ± 0.024 |
8.281 ± 0.043 | 4.832 ± 0.027 |
5.723 ± 0.045 | 7.618 ± 0.035 |
1.545 ± 0.016 | 2.647 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
