
Natrialba asiatica (strain ATCC 700177 / DSM 12278 / JCM 9576 / FERM P-10747 / NBRC 102637 / 172P1)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Natrialba; Natrialba asiatica
Average proteome isoelectric point is 4.84
Get precalculated fractions of proteins
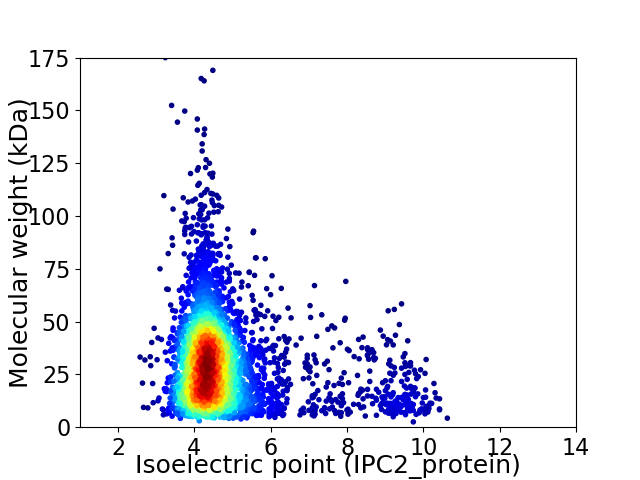
Virtual 2D-PAGE plot for 4175 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
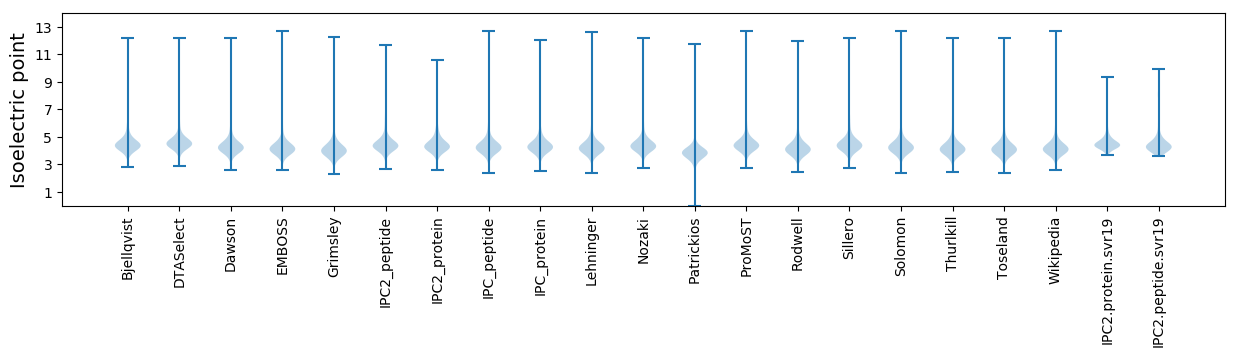
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|M0B519|M0B519_NATA1 Uncharacterized protein OS=Natrialba asiatica (strain ATCC 700177 / DSM 12278 / JCM 9576 / FERM P-10747 / NBRC 102637 / 172P1) OX=29540 GN=C481_00200 PE=4 SV=1
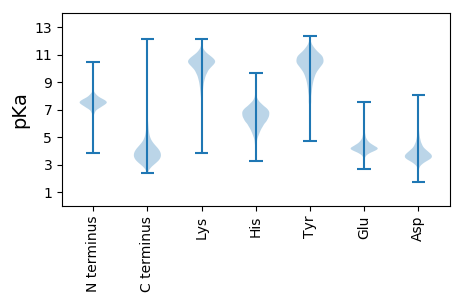
MM1 pKa = 7.64IYY3 pKa = 10.66AVTADD8 pKa = 3.35TGEE11 pKa = 4.02EE12 pKa = 3.44RR13 pKa = 11.84WRR15 pKa = 11.84FEE17 pKa = 3.71TNDD20 pKa = 3.41AVRR23 pKa = 11.84STPAVVDD30 pKa = 3.47GTLYY34 pKa = 11.0VGLHH38 pKa = 6.09DD39 pKa = 5.71QMLALDD45 pKa = 4.28AASGSEE51 pKa = 3.74QWRR54 pKa = 11.84VDD56 pKa = 3.62VAPGGSNPTVVQGEE70 pKa = 4.42SVYY73 pKa = 10.91VCRR76 pKa = 11.84QGAAFGIGGIAKK88 pKa = 9.79LDD90 pKa = 3.7SSNGDD95 pKa = 3.55VQSKK99 pKa = 9.44FATDD103 pKa = 3.31TTGGLTAAAVGDD115 pKa = 3.47QTVYY119 pKa = 10.67VGSEE123 pKa = 3.99EE124 pKa = 4.6GSLSAYY130 pKa = 10.28DD131 pKa = 4.1IEE133 pKa = 4.66TGDD136 pKa = 4.13VKK138 pKa = 10.52WSSDD142 pKa = 3.58VEE144 pKa = 4.29LGLVGPPALADD155 pKa = 3.37EE156 pKa = 4.57TVYY159 pKa = 11.11VVDD162 pKa = 4.12IDD164 pKa = 3.59ATLRR168 pKa = 11.84AFDD171 pKa = 4.68PSDD174 pKa = 3.64GTIHH178 pKa = 7.01WAEE181 pKa = 3.89QLPGVSEE188 pKa = 4.36GSPAVIGGAVYY199 pKa = 10.7LGDD202 pKa = 3.66TQGTVHH208 pKa = 7.22CLATDD213 pKa = 3.68DD214 pKa = 6.16GEE216 pKa = 4.77RR217 pKa = 11.84LWQADD222 pKa = 3.59TGNWITPSPAVTDD235 pKa = 3.57TAVYY239 pKa = 10.22VCNNDD244 pKa = 3.22GAVYY248 pKa = 10.22AIDD251 pKa = 3.51QTEE254 pKa = 4.94GEE256 pKa = 4.67ILWQFATGGQLRR268 pKa = 11.84TSPAVVDD275 pKa = 3.88GTAFVGSDD283 pKa = 3.69DD284 pKa = 3.7GSLYY288 pKa = 11.21ALAEE292 pKa = 4.36DD293 pKa = 4.13SS294 pKa = 3.86
MM1 pKa = 7.64IYY3 pKa = 10.66AVTADD8 pKa = 3.35TGEE11 pKa = 4.02EE12 pKa = 3.44RR13 pKa = 11.84WRR15 pKa = 11.84FEE17 pKa = 3.71TNDD20 pKa = 3.41AVRR23 pKa = 11.84STPAVVDD30 pKa = 3.47GTLYY34 pKa = 11.0VGLHH38 pKa = 6.09DD39 pKa = 5.71QMLALDD45 pKa = 4.28AASGSEE51 pKa = 3.74QWRR54 pKa = 11.84VDD56 pKa = 3.62VAPGGSNPTVVQGEE70 pKa = 4.42SVYY73 pKa = 10.91VCRR76 pKa = 11.84QGAAFGIGGIAKK88 pKa = 9.79LDD90 pKa = 3.7SSNGDD95 pKa = 3.55VQSKK99 pKa = 9.44FATDD103 pKa = 3.31TTGGLTAAAVGDD115 pKa = 3.47QTVYY119 pKa = 10.67VGSEE123 pKa = 3.99EE124 pKa = 4.6GSLSAYY130 pKa = 10.28DD131 pKa = 4.1IEE133 pKa = 4.66TGDD136 pKa = 4.13VKK138 pKa = 10.52WSSDD142 pKa = 3.58VEE144 pKa = 4.29LGLVGPPALADD155 pKa = 3.37EE156 pKa = 4.57TVYY159 pKa = 11.11VVDD162 pKa = 4.12IDD164 pKa = 3.59ATLRR168 pKa = 11.84AFDD171 pKa = 4.68PSDD174 pKa = 3.64GTIHH178 pKa = 7.01WAEE181 pKa = 3.89QLPGVSEE188 pKa = 4.36GSPAVIGGAVYY199 pKa = 10.7LGDD202 pKa = 3.66TQGTVHH208 pKa = 7.22CLATDD213 pKa = 3.68DD214 pKa = 6.16GEE216 pKa = 4.77RR217 pKa = 11.84LWQADD222 pKa = 3.59TGNWITPSPAVTDD235 pKa = 3.57TAVYY239 pKa = 10.22VCNNDD244 pKa = 3.22GAVYY248 pKa = 10.22AIDD251 pKa = 3.51QTEE254 pKa = 4.94GEE256 pKa = 4.67ILWQFATGGQLRR268 pKa = 11.84TSPAVVDD275 pKa = 3.88GTAFVGSDD283 pKa = 3.69DD284 pKa = 3.7GSLYY288 pKa = 11.21ALAEE292 pKa = 4.36DD293 pKa = 4.13SS294 pKa = 3.86
Molecular weight: 30.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M0B2W1|M0B2W1_NATA1 Uncharacterized protein OS=Natrialba asiatica (strain ATCC 700177 / DSM 12278 / JCM 9576 / FERM P-10747 / NBRC 102637 / 172P1) OX=29540 GN=C481_03032 PE=4 SV=1
MM1 pKa = 7.42TCFDD5 pKa = 4.15DD6 pKa = 4.78VLTKK10 pKa = 10.66RR11 pKa = 11.84LICNVRR17 pKa = 11.84MARR20 pKa = 11.84SHH22 pKa = 6.3LLKK25 pKa = 9.76TVYY28 pKa = 9.09MANLRR33 pKa = 11.84LQLEE37 pKa = 3.96QRR39 pKa = 11.84PIGWISFYY47 pKa = 11.33LSINKK52 pKa = 9.46LMIGGFVRR60 pKa = 11.84SQTALQRR67 pKa = 11.84TT68 pKa = 4.08
MM1 pKa = 7.42TCFDD5 pKa = 4.15DD6 pKa = 4.78VLTKK10 pKa = 10.66RR11 pKa = 11.84LICNVRR17 pKa = 11.84MARR20 pKa = 11.84SHH22 pKa = 6.3LLKK25 pKa = 9.76TVYY28 pKa = 9.09MANLRR33 pKa = 11.84LQLEE37 pKa = 3.96QRR39 pKa = 11.84PIGWISFYY47 pKa = 11.33LSINKK52 pKa = 9.46LMIGGFVRR60 pKa = 11.84SQTALQRR67 pKa = 11.84TT68 pKa = 4.08
Molecular weight: 7.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1209462 |
26 |
1675 |
289.7 |
31.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.68 ± 0.054 | 0.739 ± 0.012 |
8.465 ± 0.048 | 8.67 ± 0.055 |
3.309 ± 0.026 | 8.333 ± 0.04 |
2.008 ± 0.02 | 4.561 ± 0.033 |
1.682 ± 0.022 | 8.794 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.727 ± 0.015 | 2.5 ± 0.019 |
4.667 ± 0.025 | 2.598 ± 0.025 |
6.348 ± 0.041 | 5.801 ± 0.032 |
7.007 ± 0.033 | 8.308 ± 0.035 |
1.134 ± 0.015 | 2.668 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
