
bacterium I07
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
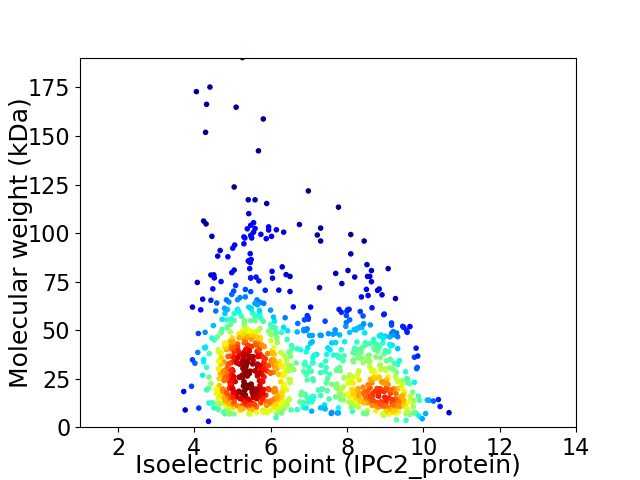
Virtual 2D-PAGE plot for 1024 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
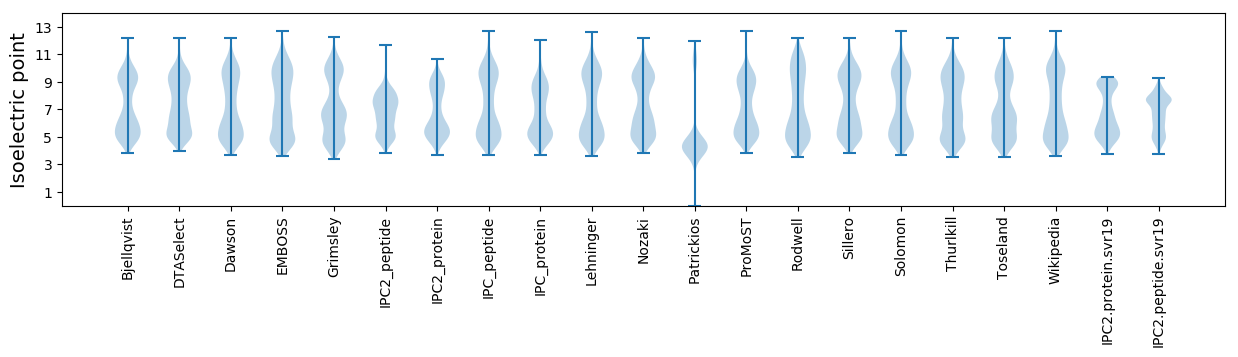
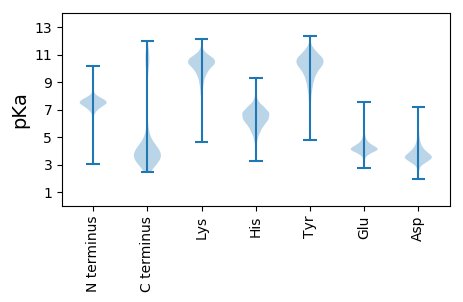
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A202DW17|A0A202DW17_9BACT B12-binding domain-containing protein OS=bacterium I07 OX=1932703 GN=BVY01_00640 PE=4 SV=1
MM1 pKa = 7.47RR2 pKa = 11.84RR3 pKa = 11.84HH4 pKa = 5.82MGLILFLFVPGLLDD18 pKa = 3.53AQYY21 pKa = 11.27GNPSLPYY28 pKa = 9.14GQGAYY33 pKa = 10.15GIQTDD38 pKa = 3.74AEE40 pKa = 4.15EE41 pKa = 5.12AEE43 pKa = 4.44EE44 pKa = 4.69GGEE47 pKa = 3.83QAYY50 pKa = 8.85TGWSEE55 pKa = 4.4SKK57 pKa = 9.47TVTITGSDD65 pKa = 3.32SGDD68 pKa = 3.2ATGYY72 pKa = 11.6ALFLDD77 pKa = 3.8IDD79 pKa = 4.26YY80 pKa = 8.38ATGMDD85 pKa = 3.81SLFRR89 pKa = 11.84DD90 pKa = 3.7LRR92 pKa = 11.84FSQDD96 pKa = 2.7GEE98 pKa = 4.13ALHH101 pKa = 6.75FWRR104 pKa = 11.84EE105 pKa = 4.18SVFKK109 pKa = 10.81SDD111 pKa = 5.3SALFWVKK118 pKa = 10.37VPSIPLGDD126 pKa = 4.11DD127 pKa = 3.37VTVTLHH133 pKa = 6.06YY134 pKa = 11.45SNDD137 pKa = 3.21TCATDD142 pKa = 3.65ISNGDD147 pKa = 3.35STFAFFDD154 pKa = 3.8DD155 pKa = 5.39FGDD158 pKa = 3.72EE159 pKa = 4.84SIDD162 pKa = 3.69SEE164 pKa = 4.22KK165 pKa = 10.58WPTSLEE171 pKa = 3.93LGQFTEE177 pKa = 4.13NANGYY182 pKa = 10.39LEE184 pKa = 4.46VNHH187 pKa = 6.72TGMITMLLQATNDD200 pKa = 3.84TVHH203 pKa = 6.83VPIAWKK209 pKa = 8.83TRR211 pKa = 11.84INQGTANLANYY222 pKa = 8.73RR223 pKa = 11.84RR224 pKa = 11.84EE225 pKa = 4.43GIGVSTGYY233 pKa = 10.91DD234 pKa = 3.12LGTTQAGTAAYY245 pKa = 9.9YY246 pKa = 10.39YY247 pKa = 10.74NHH249 pKa = 7.85DD250 pKa = 3.93DD251 pKa = 3.61ATWRR255 pKa = 11.84SDD257 pKa = 2.9EE258 pKa = 4.34STSLLAQTDD267 pKa = 4.07WVPDD271 pKa = 3.24ITFDD275 pKa = 3.18QWVVLEE281 pKa = 4.93GKK283 pKa = 10.46ASVDD287 pKa = 2.75SCYY290 pKa = 10.35FYY292 pKa = 11.2QNQAYY297 pKa = 9.99DD298 pKa = 3.73LTHH301 pKa = 7.57DD302 pKa = 4.14YY303 pKa = 11.2SPQNDD308 pKa = 3.61ADD310 pKa = 5.07LIPP313 pKa = 4.63
MM1 pKa = 7.47RR2 pKa = 11.84RR3 pKa = 11.84HH4 pKa = 5.82MGLILFLFVPGLLDD18 pKa = 3.53AQYY21 pKa = 11.27GNPSLPYY28 pKa = 9.14GQGAYY33 pKa = 10.15GIQTDD38 pKa = 3.74AEE40 pKa = 4.15EE41 pKa = 5.12AEE43 pKa = 4.44EE44 pKa = 4.69GGEE47 pKa = 3.83QAYY50 pKa = 8.85TGWSEE55 pKa = 4.4SKK57 pKa = 9.47TVTITGSDD65 pKa = 3.32SGDD68 pKa = 3.2ATGYY72 pKa = 11.6ALFLDD77 pKa = 3.8IDD79 pKa = 4.26YY80 pKa = 8.38ATGMDD85 pKa = 3.81SLFRR89 pKa = 11.84DD90 pKa = 3.7LRR92 pKa = 11.84FSQDD96 pKa = 2.7GEE98 pKa = 4.13ALHH101 pKa = 6.75FWRR104 pKa = 11.84EE105 pKa = 4.18SVFKK109 pKa = 10.81SDD111 pKa = 5.3SALFWVKK118 pKa = 10.37VPSIPLGDD126 pKa = 4.11DD127 pKa = 3.37VTVTLHH133 pKa = 6.06YY134 pKa = 11.45SNDD137 pKa = 3.21TCATDD142 pKa = 3.65ISNGDD147 pKa = 3.35STFAFFDD154 pKa = 3.8DD155 pKa = 5.39FGDD158 pKa = 3.72EE159 pKa = 4.84SIDD162 pKa = 3.69SEE164 pKa = 4.22KK165 pKa = 10.58WPTSLEE171 pKa = 3.93LGQFTEE177 pKa = 4.13NANGYY182 pKa = 10.39LEE184 pKa = 4.46VNHH187 pKa = 6.72TGMITMLLQATNDD200 pKa = 3.84TVHH203 pKa = 6.83VPIAWKK209 pKa = 8.83TRR211 pKa = 11.84INQGTANLANYY222 pKa = 8.73RR223 pKa = 11.84RR224 pKa = 11.84EE225 pKa = 4.43GIGVSTGYY233 pKa = 10.91DD234 pKa = 3.12LGTTQAGTAAYY245 pKa = 9.9YY246 pKa = 10.39YY247 pKa = 10.74NHH249 pKa = 7.85DD250 pKa = 3.93DD251 pKa = 3.61ATWRR255 pKa = 11.84SDD257 pKa = 2.9EE258 pKa = 4.34STSLLAQTDD267 pKa = 4.07WVPDD271 pKa = 3.24ITFDD275 pKa = 3.18QWVVLEE281 pKa = 4.93GKK283 pKa = 10.46ASVDD287 pKa = 2.75SCYY290 pKa = 10.35FYY292 pKa = 11.2QNQAYY297 pKa = 9.99DD298 pKa = 3.73LTHH301 pKa = 7.57DD302 pKa = 4.14YY303 pKa = 11.2SPQNDD308 pKa = 3.61ADD310 pKa = 5.07LIPP313 pKa = 4.63
Molecular weight: 34.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A202DT02|A0A202DT02_9BACT Uncharacterized protein OS=bacterium I07 OX=1932703 GN=BVY01_03535 PE=4 SV=1
MM1 pKa = 8.08LIRR4 pKa = 11.84SATSLSKK11 pKa = 10.8NSRR14 pKa = 11.84IRR16 pKa = 11.84NDD18 pKa = 2.8IFEE21 pKa = 5.73LIVTLLLTLTLLVRR35 pKa = 11.84MLVLGYY41 pKa = 10.18VSAGFSMDD49 pKa = 3.13MLGRR53 pKa = 11.84ICSFAWIEE61 pKa = 3.94KK62 pKa = 9.76QIYY65 pKa = 9.56CISCQRR71 pKa = 11.84MAIVNDD77 pKa = 4.86FGALRR82 pKa = 11.84PRR84 pKa = 11.84TQLAQLKK91 pKa = 9.76LFAFGRR97 pKa = 11.84KK98 pKa = 7.98AA99 pKa = 3.08
MM1 pKa = 8.08LIRR4 pKa = 11.84SATSLSKK11 pKa = 10.8NSRR14 pKa = 11.84IRR16 pKa = 11.84NDD18 pKa = 2.8IFEE21 pKa = 5.73LIVTLLLTLTLLVRR35 pKa = 11.84MLVLGYY41 pKa = 10.18VSAGFSMDD49 pKa = 3.13MLGRR53 pKa = 11.84ICSFAWIEE61 pKa = 3.94KK62 pKa = 9.76QIYY65 pKa = 9.56CISCQRR71 pKa = 11.84MAIVNDD77 pKa = 4.86FGALRR82 pKa = 11.84PRR84 pKa = 11.84TQLAQLKK91 pKa = 9.76LFAFGRR97 pKa = 11.84KK98 pKa = 7.98AA99 pKa = 3.08
Molecular weight: 11.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
315021 |
27 |
1666 |
307.6 |
34.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.33 ± 0.072 | 1.053 ± 0.027 |
5.852 ± 0.065 | 6.392 ± 0.061 |
4.471 ± 0.054 | 7.23 ± 0.08 |
2.265 ± 0.031 | 7.464 ± 0.074 |
6.063 ± 0.082 | 9.228 ± 0.099 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.492 ± 0.03 | 4.554 ± 0.064 |
4.272 ± 0.055 | 3.541 ± 0.041 |
5.415 ± 0.061 | 6.668 ± 0.065 |
5.166 ± 0.048 | 6.37 ± 0.065 |
1.535 ± 0.036 | 3.638 ± 0.059 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
