
gamma proteobacterium HTCC2207
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Porticoccaceae; unclassified Porticoccaceae; SAR92 clade
Average proteome isoelectric point is 5.85
Get precalculated fractions of proteins
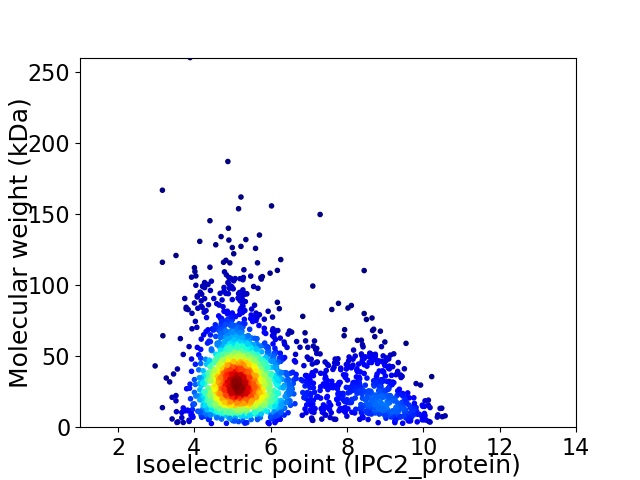
Virtual 2D-PAGE plot for 2388 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q1YR40|Q1YR40_9GAMM Gln-synt_C domain-containing protein OS=gamma proteobacterium HTCC2207 OX=314287 GN=GB2207_03989 PE=3 SV=1
MM1 pKa = 7.1THH3 pKa = 5.13TKK5 pKa = 9.45IRR7 pKa = 11.84KK8 pKa = 6.22TLISAAVAAATLSAGQLSAAQIEE31 pKa = 4.71EE32 pKa = 4.57IIVTTQKK39 pKa = 10.43RR40 pKa = 11.84QQSLQDD46 pKa = 3.43VPISVSAFSGSFIEE60 pKa = 4.85KK61 pKa = 10.33AQISDD66 pKa = 3.57AKK68 pKa = 10.25QVALLTPGVSGDD80 pKa = 3.72TDD82 pKa = 4.38DD83 pKa = 6.04SFLDD87 pKa = 3.55SMNVRR92 pKa = 11.84GISTNDD98 pKa = 3.16FGVGAEE104 pKa = 4.22PSIGLYY110 pKa = 9.88QDD112 pKa = 4.72GIYY115 pKa = 10.4LGRR118 pKa = 11.84TGGAVSSFFDD128 pKa = 3.45VEE130 pKa = 4.09MVEE133 pKa = 4.64VVKK136 pKa = 10.93GPQGTLFGRR145 pKa = 11.84NASSGAISITTAKK158 pKa = 8.57PTGEE162 pKa = 4.2AGGSIDD168 pKa = 4.98IGFGQDD174 pKa = 2.95GYY176 pKa = 11.84GEE178 pKa = 4.22VTAVINTPINDD189 pKa = 3.61QFSGRR194 pKa = 11.84LAVYY198 pKa = 9.25HH199 pKa = 5.2QQQDD203 pKa = 3.22GWVTNINDD211 pKa = 3.87GQQTGGVEE219 pKa = 3.99NTAARR224 pKa = 11.84YY225 pKa = 7.44TLAFEE230 pKa = 4.85NDD232 pKa = 4.84DD233 pKa = 3.55ITSTLMLEE241 pKa = 4.38YY242 pKa = 9.91EE243 pKa = 4.63DD244 pKa = 4.72RR245 pKa = 11.84QGPPTIYY252 pKa = 10.16QAFDD256 pKa = 3.7PDD258 pKa = 3.67EE259 pKa = 4.57TGLPFYY265 pKa = 10.56STDD268 pKa = 3.28AAEE271 pKa = 4.68DD272 pKa = 3.56QFTSDD277 pKa = 3.68VSSSDD282 pKa = 4.26LIDD285 pKa = 3.94EE286 pKa = 4.61GEE288 pKa = 4.27VWGVTLNVEE297 pKa = 3.9VDD299 pKa = 3.63LGNDD303 pKa = 3.44YY304 pKa = 9.16TLSSTTGLRR313 pKa = 11.84GHH315 pKa = 6.67NYY317 pKa = 10.31YY318 pKa = 10.79YY319 pKa = 11.06LEE321 pKa = 4.9DD322 pKa = 4.17FDD324 pKa = 5.55GGSSFLSNYY333 pKa = 8.92NQIQEE338 pKa = 3.74QDD340 pKa = 3.61YY341 pKa = 9.79FSQEE345 pKa = 3.34FRR347 pKa = 11.84INKK350 pKa = 7.0EE351 pKa = 3.79TDD353 pKa = 3.31SVSWFVGASWYY364 pKa = 10.5KK365 pKa = 10.57EE366 pKa = 3.69EE367 pKa = 4.72LKK369 pKa = 11.08VRR371 pKa = 11.84FNQTLDD377 pKa = 3.34EE378 pKa = 4.47DD379 pKa = 4.76TFCAAYY385 pKa = 8.19GTYY388 pKa = 10.12YY389 pKa = 10.44EE390 pKa = 4.77YY391 pKa = 11.32DD392 pKa = 5.31DD393 pKa = 4.12ITDD396 pKa = 3.74CATLYY401 pKa = 10.46EE402 pKa = 4.51YY403 pKa = 11.39YY404 pKa = 10.13EE405 pKa = 4.18YY406 pKa = 11.26DD407 pKa = 4.41PIVGIGTRR415 pKa = 11.84NDD417 pKa = 3.63SVDD420 pKa = 2.79VDD422 pKa = 3.84AEE424 pKa = 4.09YY425 pKa = 11.04DD426 pKa = 3.32GWGLYY431 pKa = 10.46GDD433 pKa = 3.8ATFHH437 pKa = 6.53VNEE440 pKa = 4.94DD441 pKa = 3.01LDD443 pKa = 4.7FIVGARR449 pKa = 11.84YY450 pKa = 8.09TEE452 pKa = 4.3DD453 pKa = 3.31NRR455 pKa = 11.84DD456 pKa = 3.1FAQNFGGEE464 pKa = 4.2EE465 pKa = 4.25RR466 pKa = 11.84NATWYY471 pKa = 7.53TFPFYY476 pKa = 10.64TSDD479 pKa = 3.54FVQGNDD485 pKa = 2.37KK486 pKa = 8.41WTNTSVRR493 pKa = 11.84AAVNYY498 pKa = 9.58QLSDD502 pKa = 3.59EE503 pKa = 4.37VSTYY507 pKa = 9.31ATYY510 pKa = 9.3STGYY514 pKa = 9.44KK515 pKa = 10.25AGGFNTMEE523 pKa = 4.68LSFADD528 pKa = 5.37DD529 pKa = 3.39IAVDD533 pKa = 4.15DD534 pKa = 5.23LDD536 pKa = 6.47GEE538 pKa = 4.58DD539 pKa = 5.56ALDD542 pKa = 4.29FDD544 pKa = 5.22PSLASFDD551 pKa = 4.0KK552 pKa = 11.49EE553 pKa = 3.86EE554 pKa = 4.08VTNLEE559 pKa = 4.8LGLKK563 pKa = 10.04GQFLDD568 pKa = 5.0GQMQLNAALYY578 pKa = 9.53SYY580 pKa = 10.62QFDD583 pKa = 4.03GMQSGYY589 pKa = 10.59YY590 pKa = 9.46VDD592 pKa = 4.26GRR594 pKa = 11.84YY595 pKa = 7.16TVANVGDD602 pKa = 4.0AEE604 pKa = 4.49GSGLEE609 pKa = 3.96LDD611 pKa = 3.96MRR613 pKa = 11.84YY614 pKa = 10.11LPTDD618 pKa = 3.01NVDD621 pKa = 3.21IYY623 pKa = 11.4LGLAWADD630 pKa = 3.75SEE632 pKa = 4.4LTKK635 pKa = 10.78PNSSLSADD643 pKa = 3.69FCEE646 pKa = 4.97DD647 pKa = 3.45DD648 pKa = 4.03CTGAMLPGTVDD659 pKa = 4.08FSAALVATYY668 pKa = 10.29SRR670 pKa = 11.84PIEE673 pKa = 4.57GGDD676 pKa = 4.68LNLTWEE682 pKa = 4.51TFHH685 pKa = 7.33QGASPGFGDD694 pKa = 5.1FSRR697 pKa = 11.84EE698 pKa = 3.76PIMLDD703 pKa = 2.86EE704 pKa = 4.51FTVSNLRR711 pKa = 11.84VGYY714 pKa = 10.18DD715 pKa = 3.38SSDD718 pKa = 3.16NWSMTVWVNNVFDD731 pKa = 3.47TFYY734 pKa = 10.77YY735 pKa = 10.36KK736 pKa = 10.68GVAPADD742 pKa = 4.51GIIAPHH748 pKa = 5.97YY749 pKa = 10.28FGFSEE754 pKa = 4.29PRR756 pKa = 11.84RR757 pKa = 11.84MGLDD761 pKa = 3.5LAYY764 pKa = 9.92KK765 pKa = 9.96FF766 pKa = 4.42
MM1 pKa = 7.1THH3 pKa = 5.13TKK5 pKa = 9.45IRR7 pKa = 11.84KK8 pKa = 6.22TLISAAVAAATLSAGQLSAAQIEE31 pKa = 4.71EE32 pKa = 4.57IIVTTQKK39 pKa = 10.43RR40 pKa = 11.84QQSLQDD46 pKa = 3.43VPISVSAFSGSFIEE60 pKa = 4.85KK61 pKa = 10.33AQISDD66 pKa = 3.57AKK68 pKa = 10.25QVALLTPGVSGDD80 pKa = 3.72TDD82 pKa = 4.38DD83 pKa = 6.04SFLDD87 pKa = 3.55SMNVRR92 pKa = 11.84GISTNDD98 pKa = 3.16FGVGAEE104 pKa = 4.22PSIGLYY110 pKa = 9.88QDD112 pKa = 4.72GIYY115 pKa = 10.4LGRR118 pKa = 11.84TGGAVSSFFDD128 pKa = 3.45VEE130 pKa = 4.09MVEE133 pKa = 4.64VVKK136 pKa = 10.93GPQGTLFGRR145 pKa = 11.84NASSGAISITTAKK158 pKa = 8.57PTGEE162 pKa = 4.2AGGSIDD168 pKa = 4.98IGFGQDD174 pKa = 2.95GYY176 pKa = 11.84GEE178 pKa = 4.22VTAVINTPINDD189 pKa = 3.61QFSGRR194 pKa = 11.84LAVYY198 pKa = 9.25HH199 pKa = 5.2QQQDD203 pKa = 3.22GWVTNINDD211 pKa = 3.87GQQTGGVEE219 pKa = 3.99NTAARR224 pKa = 11.84YY225 pKa = 7.44TLAFEE230 pKa = 4.85NDD232 pKa = 4.84DD233 pKa = 3.55ITSTLMLEE241 pKa = 4.38YY242 pKa = 9.91EE243 pKa = 4.63DD244 pKa = 4.72RR245 pKa = 11.84QGPPTIYY252 pKa = 10.16QAFDD256 pKa = 3.7PDD258 pKa = 3.67EE259 pKa = 4.57TGLPFYY265 pKa = 10.56STDD268 pKa = 3.28AAEE271 pKa = 4.68DD272 pKa = 3.56QFTSDD277 pKa = 3.68VSSSDD282 pKa = 4.26LIDD285 pKa = 3.94EE286 pKa = 4.61GEE288 pKa = 4.27VWGVTLNVEE297 pKa = 3.9VDD299 pKa = 3.63LGNDD303 pKa = 3.44YY304 pKa = 9.16TLSSTTGLRR313 pKa = 11.84GHH315 pKa = 6.67NYY317 pKa = 10.31YY318 pKa = 10.79YY319 pKa = 11.06LEE321 pKa = 4.9DD322 pKa = 4.17FDD324 pKa = 5.55GGSSFLSNYY333 pKa = 8.92NQIQEE338 pKa = 3.74QDD340 pKa = 3.61YY341 pKa = 9.79FSQEE345 pKa = 3.34FRR347 pKa = 11.84INKK350 pKa = 7.0EE351 pKa = 3.79TDD353 pKa = 3.31SVSWFVGASWYY364 pKa = 10.5KK365 pKa = 10.57EE366 pKa = 3.69EE367 pKa = 4.72LKK369 pKa = 11.08VRR371 pKa = 11.84FNQTLDD377 pKa = 3.34EE378 pKa = 4.47DD379 pKa = 4.76TFCAAYY385 pKa = 8.19GTYY388 pKa = 10.12YY389 pKa = 10.44EE390 pKa = 4.77YY391 pKa = 11.32DD392 pKa = 5.31DD393 pKa = 4.12ITDD396 pKa = 3.74CATLYY401 pKa = 10.46EE402 pKa = 4.51YY403 pKa = 11.39YY404 pKa = 10.13EE405 pKa = 4.18YY406 pKa = 11.26DD407 pKa = 4.41PIVGIGTRR415 pKa = 11.84NDD417 pKa = 3.63SVDD420 pKa = 2.79VDD422 pKa = 3.84AEE424 pKa = 4.09YY425 pKa = 11.04DD426 pKa = 3.32GWGLYY431 pKa = 10.46GDD433 pKa = 3.8ATFHH437 pKa = 6.53VNEE440 pKa = 4.94DD441 pKa = 3.01LDD443 pKa = 4.7FIVGARR449 pKa = 11.84YY450 pKa = 8.09TEE452 pKa = 4.3DD453 pKa = 3.31NRR455 pKa = 11.84DD456 pKa = 3.1FAQNFGGEE464 pKa = 4.2EE465 pKa = 4.25RR466 pKa = 11.84NATWYY471 pKa = 7.53TFPFYY476 pKa = 10.64TSDD479 pKa = 3.54FVQGNDD485 pKa = 2.37KK486 pKa = 8.41WTNTSVRR493 pKa = 11.84AAVNYY498 pKa = 9.58QLSDD502 pKa = 3.59EE503 pKa = 4.37VSTYY507 pKa = 9.31ATYY510 pKa = 9.3STGYY514 pKa = 9.44KK515 pKa = 10.25AGGFNTMEE523 pKa = 4.68LSFADD528 pKa = 5.37DD529 pKa = 3.39IAVDD533 pKa = 4.15DD534 pKa = 5.23LDD536 pKa = 6.47GEE538 pKa = 4.58DD539 pKa = 5.56ALDD542 pKa = 4.29FDD544 pKa = 5.22PSLASFDD551 pKa = 4.0KK552 pKa = 11.49EE553 pKa = 3.86EE554 pKa = 4.08VTNLEE559 pKa = 4.8LGLKK563 pKa = 10.04GQFLDD568 pKa = 5.0GQMQLNAALYY578 pKa = 9.53SYY580 pKa = 10.62QFDD583 pKa = 4.03GMQSGYY589 pKa = 10.59YY590 pKa = 9.46VDD592 pKa = 4.26GRR594 pKa = 11.84YY595 pKa = 7.16TVANVGDD602 pKa = 4.0AEE604 pKa = 4.49GSGLEE609 pKa = 3.96LDD611 pKa = 3.96MRR613 pKa = 11.84YY614 pKa = 10.11LPTDD618 pKa = 3.01NVDD621 pKa = 3.21IYY623 pKa = 11.4LGLAWADD630 pKa = 3.75SEE632 pKa = 4.4LTKK635 pKa = 10.78PNSSLSADD643 pKa = 3.69FCEE646 pKa = 4.97DD647 pKa = 3.45DD648 pKa = 4.03CTGAMLPGTVDD659 pKa = 4.08FSAALVATYY668 pKa = 10.29SRR670 pKa = 11.84PIEE673 pKa = 4.57GGDD676 pKa = 4.68LNLTWEE682 pKa = 4.51TFHH685 pKa = 7.33QGASPGFGDD694 pKa = 5.1FSRR697 pKa = 11.84EE698 pKa = 3.76PIMLDD703 pKa = 2.86EE704 pKa = 4.51FTVSNLRR711 pKa = 11.84VGYY714 pKa = 10.18DD715 pKa = 3.38SSDD718 pKa = 3.16NWSMTVWVNNVFDD731 pKa = 3.47TFYY734 pKa = 10.77YY735 pKa = 10.36KK736 pKa = 10.68GVAPADD742 pKa = 4.51GIIAPHH748 pKa = 5.97YY749 pKa = 10.28FGFSEE754 pKa = 4.29PRR756 pKa = 11.84RR757 pKa = 11.84MGLDD761 pKa = 3.5LAYY764 pKa = 9.92KK765 pKa = 9.96FF766 pKa = 4.42
Molecular weight: 84.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q1YV64|Q1YV64_9GAMM Flagellar motor switch protein FliG OS=gamma proteobacterium HTCC2207 OX=314287 GN=GB2207_08546 PE=3 SV=1
MM1 pKa = 7.86PKK3 pKa = 7.87THH5 pKa = 7.07KK6 pKa = 10.45AFHH9 pKa = 6.06HH10 pKa = 6.34TPHH13 pKa = 6.36KK14 pKa = 9.15TGDD17 pKa = 3.72YY18 pKa = 10.38AALAHH23 pKa = 6.43FSDD26 pKa = 4.03RR27 pKa = 11.84ATWRR31 pKa = 11.84QSTVARR37 pKa = 11.84THH39 pKa = 5.95SVPAFWRR46 pKa = 11.84DD47 pKa = 3.11WLLHH51 pKa = 6.29PGSLTQRR58 pKa = 11.84LLDD61 pKa = 4.16ASDD64 pKa = 3.72GQFKK68 pKa = 11.05VGVLSQSLLRR78 pKa = 11.84PSLSEE83 pKa = 3.82RR84 pKa = 11.84RR85 pKa = 11.84ALSLPDD91 pKa = 3.35HH92 pKa = 6.77RR93 pKa = 11.84LALVRR98 pKa = 11.84EE99 pKa = 4.43VILIGRR105 pKa = 11.84GVPWVFARR113 pKa = 11.84SVIPLQTLTGRR124 pKa = 11.84LRR126 pKa = 11.84KK127 pKa = 9.41LRR129 pKa = 11.84RR130 pKa = 11.84LDD132 pKa = 3.6SRR134 pKa = 11.84PLGALLFSDD143 pKa = 3.44STMTRR148 pKa = 11.84EE149 pKa = 3.95PLEE152 pKa = 4.16WACIEE157 pKa = 4.62PNGQPLAAEE166 pKa = 4.82LEE168 pKa = 4.43AMDD171 pKa = 3.9KK172 pKa = 10.48PVWGRR177 pKa = 11.84RR178 pKa = 11.84SVFKK182 pKa = 10.86LSAKK186 pKa = 9.98PMLVCEE192 pKa = 4.24VFLPSFSLQDD202 pKa = 3.28VANNLTRR209 pKa = 11.84SS210 pKa = 3.5
MM1 pKa = 7.86PKK3 pKa = 7.87THH5 pKa = 7.07KK6 pKa = 10.45AFHH9 pKa = 6.06HH10 pKa = 6.34TPHH13 pKa = 6.36KK14 pKa = 9.15TGDD17 pKa = 3.72YY18 pKa = 10.38AALAHH23 pKa = 6.43FSDD26 pKa = 4.03RR27 pKa = 11.84ATWRR31 pKa = 11.84QSTVARR37 pKa = 11.84THH39 pKa = 5.95SVPAFWRR46 pKa = 11.84DD47 pKa = 3.11WLLHH51 pKa = 6.29PGSLTQRR58 pKa = 11.84LLDD61 pKa = 4.16ASDD64 pKa = 3.72GQFKK68 pKa = 11.05VGVLSQSLLRR78 pKa = 11.84PSLSEE83 pKa = 3.82RR84 pKa = 11.84RR85 pKa = 11.84ALSLPDD91 pKa = 3.35HH92 pKa = 6.77RR93 pKa = 11.84LALVRR98 pKa = 11.84EE99 pKa = 4.43VILIGRR105 pKa = 11.84GVPWVFARR113 pKa = 11.84SVIPLQTLTGRR124 pKa = 11.84LRR126 pKa = 11.84KK127 pKa = 9.41LRR129 pKa = 11.84RR130 pKa = 11.84LDD132 pKa = 3.6SRR134 pKa = 11.84PLGALLFSDD143 pKa = 3.44STMTRR148 pKa = 11.84EE149 pKa = 3.95PLEE152 pKa = 4.16WACIEE157 pKa = 4.62PNGQPLAAEE166 pKa = 4.82LEE168 pKa = 4.43AMDD171 pKa = 3.9KK172 pKa = 10.48PVWGRR177 pKa = 11.84RR178 pKa = 11.84SVFKK182 pKa = 10.86LSAKK186 pKa = 9.98PMLVCEE192 pKa = 4.24VFLPSFSLQDD202 pKa = 3.28VANNLTRR209 pKa = 11.84SS210 pKa = 3.5
Molecular weight: 23.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
778587 |
21 |
2403 |
326.0 |
35.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.686 ± 0.053 | 1.015 ± 0.019 |
5.938 ± 0.037 | 6.067 ± 0.042 |
3.903 ± 0.032 | 7.623 ± 0.051 |
1.982 ± 0.022 | 6.157 ± 0.036 |
4.298 ± 0.036 | 10.258 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.612 ± 0.026 | 3.835 ± 0.031 |
4.15 ± 0.028 | 4.339 ± 0.037 |
5.036 ± 0.04 | 6.851 ± 0.047 |
5.197 ± 0.035 | 7.055 ± 0.043 |
1.235 ± 0.018 | 2.762 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
