
Rotavirus G pigeon/HK18
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Rotavirus; Rotavirus G
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
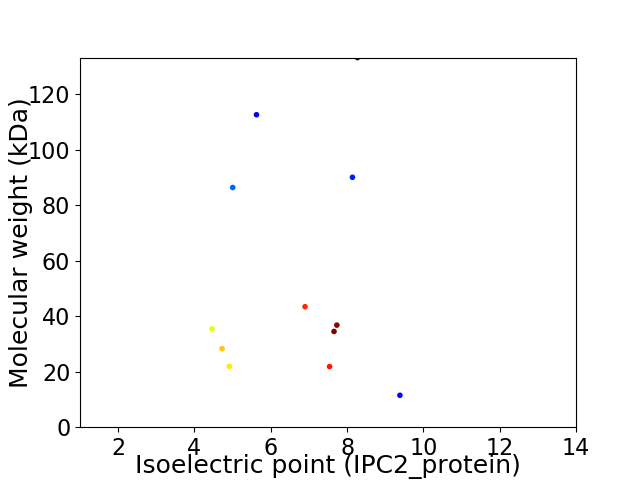
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
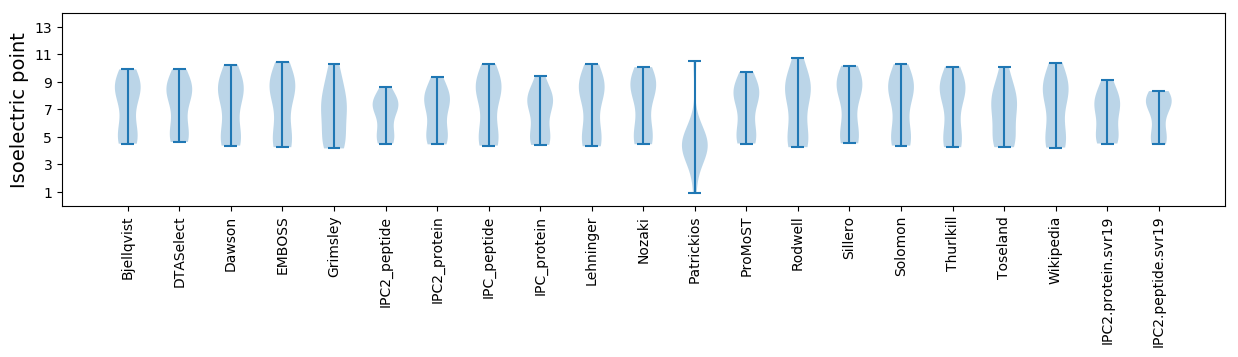
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U3R2P5|U3R2P5_9REOV VP3 OS=Rotavirus G pigeon/HK18 OX=1399970 PE=4 SV=1
MM1 pKa = 7.92AEE3 pKa = 3.97LLKK6 pKa = 11.13VCFAEE11 pKa = 4.31MMKK14 pKa = 10.41SGNCSFEE21 pKa = 4.4SFMEE25 pKa = 4.12KK26 pKa = 9.52MDD28 pKa = 4.31DD29 pKa = 3.63AGINVDD35 pKa = 3.42AWKK38 pKa = 9.99QAYY41 pKa = 9.64DD42 pKa = 3.66GNKK45 pKa = 9.57LPKK48 pKa = 10.34RR49 pKa = 11.84MTKK52 pKa = 8.6STLAIQNSNLEE63 pKa = 4.16KK64 pKa = 10.65EE65 pKa = 4.48IVTLRR70 pKa = 11.84TKK72 pKa = 7.46EE73 pKa = 3.79HH74 pKa = 6.43RR75 pKa = 11.84KK76 pKa = 10.48GYY78 pKa = 10.38DD79 pKa = 2.91KK80 pKa = 11.36NKK82 pKa = 8.78RR83 pKa = 11.84TLANFDD89 pKa = 3.56VGKK92 pKa = 10.94KK93 pKa = 10.03NGNTVLIPKK102 pKa = 8.57TRR104 pKa = 11.84LAEE107 pKa = 4.73IILLNSSHH115 pKa = 6.9DD116 pKa = 4.39LKK118 pKa = 11.33LSAPPSDD125 pKa = 3.76YY126 pKa = 10.65IEE128 pKa = 4.19YY129 pKa = 11.0LEE131 pKa = 5.74DD132 pKa = 3.56EE133 pKa = 4.13NSKK136 pKa = 10.95LKK138 pKa = 10.82EE139 pKa = 3.87EE140 pKa = 4.72NFNLMQNNHH149 pKa = 5.64YY150 pKa = 10.59LYY152 pKa = 9.78NQYY155 pKa = 10.55HH156 pKa = 6.59ALHH159 pKa = 5.93VLSYY163 pKa = 11.16GLQEE167 pKa = 3.95TNNWQKK173 pKa = 10.93EE174 pKa = 4.52VIQRR178 pKa = 11.84MQEE181 pKa = 3.9NEE183 pKa = 3.94FEE185 pKa = 4.24MSSEE189 pKa = 3.86INDD192 pKa = 3.69LKK194 pKa = 11.26SVVRR198 pKa = 11.84TLSRR202 pKa = 11.84RR203 pKa = 11.84LNYY206 pKa = 10.54DD207 pKa = 2.59VDD209 pKa = 4.3FDD211 pKa = 4.24EE212 pKa = 6.39EE213 pKa = 4.57SDD215 pKa = 3.3QGYY218 pKa = 10.1SSDD221 pKa = 4.62VIIEE225 pKa = 3.98SDD227 pKa = 3.44QEE229 pKa = 4.35DD230 pKa = 4.45NINVDD235 pKa = 3.47EE236 pKa = 4.58NDD238 pKa = 3.64NNEE241 pKa = 4.39HH242 pKa = 7.32DD243 pKa = 3.97SSADD247 pKa = 3.31EE248 pKa = 4.17NEE250 pKa = 4.31IIEE253 pKa = 5.06DD254 pKa = 4.07LDD256 pKa = 4.29DD257 pKa = 5.24LLEE260 pKa = 4.28QYY262 pKa = 7.21EE263 pKa = 4.19QRR265 pKa = 11.84QRR267 pKa = 11.84MEE269 pKa = 3.84MARR272 pKa = 11.84NALVEE277 pKa = 4.12LQIQDD282 pKa = 4.7DD283 pKa = 4.38EE284 pKa = 4.91NSDD287 pKa = 3.8DD288 pKa = 3.61EE289 pKa = 5.61DD290 pKa = 4.99YY291 pKa = 11.63NFLYY295 pKa = 10.54FGEE298 pKa = 4.5RR299 pKa = 11.84EE300 pKa = 4.12HH301 pKa = 7.34RR302 pKa = 3.8
MM1 pKa = 7.92AEE3 pKa = 3.97LLKK6 pKa = 11.13VCFAEE11 pKa = 4.31MMKK14 pKa = 10.41SGNCSFEE21 pKa = 4.4SFMEE25 pKa = 4.12KK26 pKa = 9.52MDD28 pKa = 4.31DD29 pKa = 3.63AGINVDD35 pKa = 3.42AWKK38 pKa = 9.99QAYY41 pKa = 9.64DD42 pKa = 3.66GNKK45 pKa = 9.57LPKK48 pKa = 10.34RR49 pKa = 11.84MTKK52 pKa = 8.6STLAIQNSNLEE63 pKa = 4.16KK64 pKa = 10.65EE65 pKa = 4.48IVTLRR70 pKa = 11.84TKK72 pKa = 7.46EE73 pKa = 3.79HH74 pKa = 6.43RR75 pKa = 11.84KK76 pKa = 10.48GYY78 pKa = 10.38DD79 pKa = 2.91KK80 pKa = 11.36NKK82 pKa = 8.78RR83 pKa = 11.84TLANFDD89 pKa = 3.56VGKK92 pKa = 10.94KK93 pKa = 10.03NGNTVLIPKK102 pKa = 8.57TRR104 pKa = 11.84LAEE107 pKa = 4.73IILLNSSHH115 pKa = 6.9DD116 pKa = 4.39LKK118 pKa = 11.33LSAPPSDD125 pKa = 3.76YY126 pKa = 10.65IEE128 pKa = 4.19YY129 pKa = 11.0LEE131 pKa = 5.74DD132 pKa = 3.56EE133 pKa = 4.13NSKK136 pKa = 10.95LKK138 pKa = 10.82EE139 pKa = 3.87EE140 pKa = 4.72NFNLMQNNHH149 pKa = 5.64YY150 pKa = 10.59LYY152 pKa = 9.78NQYY155 pKa = 10.55HH156 pKa = 6.59ALHH159 pKa = 5.93VLSYY163 pKa = 11.16GLQEE167 pKa = 3.95TNNWQKK173 pKa = 10.93EE174 pKa = 4.52VIQRR178 pKa = 11.84MQEE181 pKa = 3.9NEE183 pKa = 3.94FEE185 pKa = 4.24MSSEE189 pKa = 3.86INDD192 pKa = 3.69LKK194 pKa = 11.26SVVRR198 pKa = 11.84TLSRR202 pKa = 11.84RR203 pKa = 11.84LNYY206 pKa = 10.54DD207 pKa = 2.59VDD209 pKa = 4.3FDD211 pKa = 4.24EE212 pKa = 6.39EE213 pKa = 4.57SDD215 pKa = 3.3QGYY218 pKa = 10.1SSDD221 pKa = 4.62VIIEE225 pKa = 3.98SDD227 pKa = 3.44QEE229 pKa = 4.35DD230 pKa = 4.45NINVDD235 pKa = 3.47EE236 pKa = 4.58NDD238 pKa = 3.64NNEE241 pKa = 4.39HH242 pKa = 7.32DD243 pKa = 3.97SSADD247 pKa = 3.31EE248 pKa = 4.17NEE250 pKa = 4.31IIEE253 pKa = 5.06DD254 pKa = 4.07LDD256 pKa = 4.29DD257 pKa = 5.24LLEE260 pKa = 4.28QYY262 pKa = 7.21EE263 pKa = 4.19QRR265 pKa = 11.84QRR267 pKa = 11.84MEE269 pKa = 3.84MARR272 pKa = 11.84NALVEE277 pKa = 4.12LQIQDD282 pKa = 4.7DD283 pKa = 4.38EE284 pKa = 4.91NSDD287 pKa = 3.8DD288 pKa = 3.61EE289 pKa = 5.61DD290 pKa = 4.99YY291 pKa = 11.63NFLYY295 pKa = 10.54FGEE298 pKa = 4.5RR299 pKa = 11.84EE300 pKa = 4.12HH301 pKa = 7.34RR302 pKa = 3.8
Molecular weight: 35.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U3QY89|U3QY89_9REOV RNA-directed RNA polymerase OS=Rotavirus G pigeon/HK18 OX=1399970 PE=3 SV=1
MM1 pKa = 7.69GSHH4 pKa = 5.48QSSYY8 pKa = 9.5QVNNQNTIISNSKK21 pKa = 10.09NLDD24 pKa = 3.72FKK26 pKa = 11.06PSTSSALSQVNLFFVIGAAVGVFLLTALIVSVILNIYY63 pKa = 8.84LCRR66 pKa = 11.84RR67 pKa = 11.84VKK69 pKa = 10.56NGKK72 pKa = 9.34KK73 pKa = 10.31YY74 pKa = 10.42NGRR77 pKa = 11.84TNFQSGTTSRR87 pKa = 11.84HH88 pKa = 3.6NTKK91 pKa = 9.55MDD93 pKa = 3.39EE94 pKa = 4.25HH95 pKa = 6.54EE96 pKa = 4.75HH97 pKa = 5.84LQSSANVV104 pKa = 3.3
MM1 pKa = 7.69GSHH4 pKa = 5.48QSSYY8 pKa = 9.5QVNNQNTIISNSKK21 pKa = 10.09NLDD24 pKa = 3.72FKK26 pKa = 11.06PSTSSALSQVNLFFVIGAAVGVFLLTALIVSVILNIYY63 pKa = 8.84LCRR66 pKa = 11.84RR67 pKa = 11.84VKK69 pKa = 10.56NGKK72 pKa = 9.34KK73 pKa = 10.31YY74 pKa = 10.42NGRR77 pKa = 11.84TNFQSGTTSRR87 pKa = 11.84HH88 pKa = 3.6NTKK91 pKa = 9.55MDD93 pKa = 3.39EE94 pKa = 4.25HH95 pKa = 6.54EE96 pKa = 4.75HH97 pKa = 5.84LQSSANVV104 pKa = 3.3
Molecular weight: 11.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5701 |
104 |
1160 |
475.1 |
54.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.683 ± 0.457 | 1.07 ± 0.277 |
6.683 ± 0.494 | 6.841 ± 0.576 |
3.894 ± 0.36 | 3.684 ± 0.231 |
1.561 ± 0.203 | 8.314 ± 0.318 |
7.823 ± 0.537 | 7.806 ± 0.556 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.245 ± 0.234 | 6.806 ± 0.574 |
3.052 ± 0.354 | 3.982 ± 0.254 |
5.087 ± 0.311 | 6.999 ± 0.375 |
6.016 ± 0.39 | 6.209 ± 0.28 |
0.772 ± 0.13 | 4.455 ± 0.518 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
