
Ectothiorhodospira mobilis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales;
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
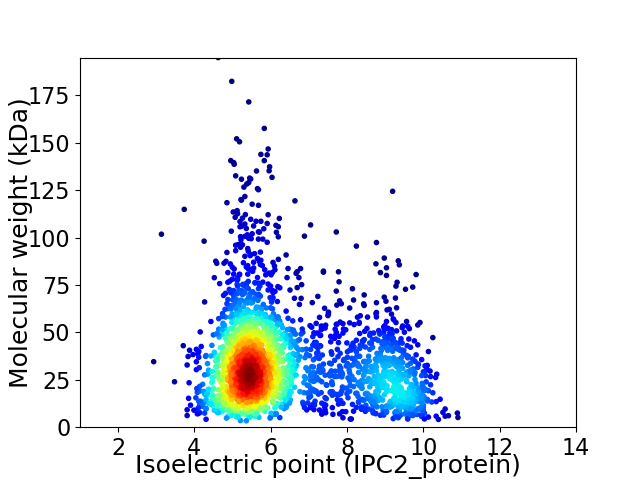
Virtual 2D-PAGE plot for 2413 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I4T0K3|A0A1I4T0K3_ECTMO Type IV pilus assembly protein PilQ OS=Ectothiorhodospira mobilis OX=195064 GN=SAMN05421721_12613 PE=4 SV=1
MM1 pKa = 7.52NRR3 pKa = 11.84FLSFALAVMTALIPAYY19 pKa = 9.82IMAEE23 pKa = 4.02PLDD26 pKa = 4.39LADD29 pKa = 5.36APLATATSAEE39 pKa = 4.41PNVMLLIDD47 pKa = 4.11TSGSMDD53 pKa = 3.78NIIWAEE59 pKa = 4.19GYY61 pKa = 10.03DD62 pKa = 3.84PSVTYY67 pKa = 10.29PGGFDD72 pKa = 4.39ANDD75 pKa = 3.91GNIHH79 pKa = 7.16LYY81 pKa = 8.11ATKK84 pKa = 10.82DD85 pKa = 3.4FGNCGNNGEE94 pKa = 4.25WVEE97 pKa = 4.6DD98 pKa = 3.34QGGWKK103 pKa = 9.74DD104 pKa = 3.62LCLPDD109 pKa = 4.8PVGGDD114 pKa = 3.17ATRR117 pKa = 11.84YY118 pKa = 6.87TGNYY122 pKa = 9.36LNYY125 pKa = 10.17LFYY128 pKa = 10.3IYY130 pKa = 10.58TEE132 pKa = 4.95DD133 pKa = 4.95DD134 pKa = 3.21TDD136 pKa = 3.99LTDD139 pKa = 3.52GTIPTEE145 pKa = 3.82TRR147 pKa = 11.84MQVAKK152 pKa = 10.16EE153 pKa = 3.97VASQLVEE160 pKa = 3.95NTPGMRR166 pKa = 11.84FGVSQFYY173 pKa = 10.75GPTWDD178 pKa = 3.37DD179 pKa = 3.56HH180 pKa = 6.38RR181 pKa = 11.84WGEE184 pKa = 4.09GAEE187 pKa = 4.21VVAACGSSVSDD198 pKa = 3.78LTSEE202 pKa = 4.06IAGFTASTNTPLAEE216 pKa = 4.03ALYY219 pKa = 10.32EE220 pKa = 3.59ITRR223 pKa = 11.84YY224 pKa = 8.95YY225 pKa = 10.91RR226 pKa = 11.84GMDD229 pKa = 3.13AYY231 pKa = 9.62YY232 pKa = 10.53TNDD235 pKa = 3.96DD236 pKa = 3.76FASPIQYY243 pKa = 9.84RR244 pKa = 11.84CQKK247 pKa = 9.8NFTVVISDD255 pKa = 4.11GLPTYY260 pKa = 7.93DD261 pKa = 3.94TNIPGNDD268 pKa = 3.51PAEE271 pKa = 4.4SSGRR275 pKa = 11.84ALPDD279 pKa = 3.2WDD281 pKa = 4.32GLSPDD286 pKa = 4.71TDD288 pKa = 3.76PADD291 pKa = 3.85YY292 pKa = 10.56PDD294 pKa = 4.35FPQYY298 pKa = 11.31SDD300 pKa = 3.83GFQPDD305 pKa = 4.95GYY307 pKa = 9.23PSEE310 pKa = 4.5EE311 pKa = 4.75GYY313 pKa = 10.92SLYY316 pKa = 10.89LDD318 pKa = 5.87DD319 pKa = 4.46IAKK322 pKa = 10.25FGWDD326 pKa = 2.73IDD328 pKa = 3.84LRR330 pKa = 11.84TGGTDD335 pKa = 3.34DD336 pKa = 5.15AGMSFDD342 pKa = 5.59DD343 pKa = 5.17PAFEE347 pKa = 4.0QQNMGTYY354 pKa = 9.99TIGFSTANQMLEE366 pKa = 3.78DD367 pKa = 3.72AAEE370 pKa = 4.32YY371 pKa = 10.79GHH373 pKa = 7.04GEE375 pKa = 4.33YY376 pKa = 9.57LTAANAAEE384 pKa = 4.61LSAALQAAISDD395 pKa = 3.48IAARR399 pKa = 11.84SSSSASATSNTGYY412 pKa = 9.81IASGSRR418 pKa = 11.84VFQARR423 pKa = 11.84YY424 pKa = 9.91NSSNWTGQLLAFAIEE439 pKa = 4.23SDD441 pKa = 3.94VDD443 pKa = 3.86SEE445 pKa = 5.77DD446 pKa = 3.57YY447 pKa = 11.46GFLQRR452 pKa = 11.84NGPVTEE458 pKa = 4.1GAVWDD463 pKa = 3.95AAEE466 pKa = 5.06EE467 pKa = 4.11IPAAGSRR474 pKa = 11.84TIMTYY479 pKa = 10.69NGGGVPFAWTDD490 pKa = 3.31LSTAQQSALGSEE502 pKa = 4.75DD503 pKa = 3.19VLSYY507 pKa = 11.22LRR509 pKa = 11.84GDD511 pKa = 3.46QSQEE515 pKa = 3.72VSSGGSFRR523 pKa = 11.84NRR525 pKa = 11.84DD526 pKa = 3.58ALLGDD531 pKa = 4.06IVNSNPVFMGAPPFNYY547 pKa = 9.52PDD549 pKa = 3.84DD550 pKa = 4.26WNGDD554 pKa = 3.45TTEE557 pKa = 5.02PEE559 pKa = 4.01DD560 pKa = 3.82ASPYY564 pKa = 10.79SSFAVNQNDD573 pKa = 4.5RR574 pKa = 11.84TPMIYY579 pKa = 10.3VGANDD584 pKa = 3.79GMLHH588 pKa = 6.42GFDD591 pKa = 4.66AEE593 pKa = 4.38TGEE596 pKa = 4.08EE597 pKa = 3.79RR598 pKa = 11.84MAYY601 pKa = 9.96VPGSVFEE608 pKa = 4.57NLDD611 pKa = 3.57EE612 pKa = 5.82LVDD615 pKa = 3.13TDD617 pKa = 3.68YY618 pKa = 11.72AHH620 pKa = 7.85RR621 pKa = 11.84YY622 pKa = 8.26FVDD625 pKa = 4.88GSPTIADD632 pKa = 3.75AYY634 pKa = 11.09FDD636 pKa = 4.61GSWHH640 pKa = 5.61TVLLGGLNKK649 pKa = 10.28GGQGIYY655 pKa = 10.77ALDD658 pKa = 3.48VTTATFSEE666 pKa = 4.89GNADD670 pKa = 4.04STVLWEE676 pKa = 4.38FTDD679 pKa = 4.63ADD681 pKa = 4.42DD682 pKa = 4.67VDD684 pKa = 4.04LGYY687 pKa = 9.28TFSQPNIVRR696 pKa = 11.84LHH698 pKa = 5.72NGQWAAIFGNGYY710 pKa = 10.24NNTRR714 pKa = 11.84ADD716 pKa = 3.7GDD718 pKa = 3.67ASTTGHH724 pKa = 6.12AVLYY728 pKa = 9.9IVSIEE733 pKa = 4.21DD734 pKa = 3.12GSLIRR739 pKa = 11.84KK740 pKa = 8.99IDD742 pKa = 3.57TGAGTATTPNGLATVSPVDD761 pKa = 3.19VDD763 pKa = 2.83GDD765 pKa = 4.35YY766 pKa = 11.1IVDD769 pKa = 3.54RR770 pKa = 11.84VYY772 pKa = 11.24AGDD775 pKa = 3.58LQGNLWRR782 pKa = 11.84FDD784 pKa = 4.29LDD786 pKa = 4.97DD787 pKa = 4.83GNPNQWGIPFSQGQTHH803 pKa = 6.41EE804 pKa = 4.61PLFSACSGTPCDD816 pKa = 5.01SNRR819 pKa = 11.84QPITVRR825 pKa = 11.84PEE827 pKa = 3.28VGRR830 pKa = 11.84APNKK834 pKa = 9.46PGAVMLYY841 pKa = 10.04FGTGKK846 pKa = 10.44YY847 pKa = 10.33LEE849 pKa = 4.75NGDD852 pKa = 4.25GNVADD857 pKa = 4.41TPQMQRR863 pKa = 11.84FYY865 pKa = 11.59GVLDD869 pKa = 3.64EE870 pKa = 6.03DD871 pKa = 4.72LPANQFDD878 pKa = 4.14TLVPGDD884 pKa = 4.42LLEE887 pKa = 4.17QEE889 pKa = 4.68ILVEE893 pKa = 4.13SSFNVEE899 pKa = 4.37GNSHH903 pKa = 5.63SLRR906 pKa = 11.84VTSDD910 pKa = 3.27YY911 pKa = 11.52TLGSEE916 pKa = 4.68DD917 pKa = 3.68KK918 pKa = 10.83GWYY921 pKa = 8.96IDD923 pKa = 4.05LAPPGTGSVGEE934 pKa = 4.25IAVTAPLLRR943 pKa = 11.84AGRR946 pKa = 11.84VFFTTLIPGEE956 pKa = 4.88DD957 pKa = 3.53PCQPGGQGWLMEE969 pKa = 4.68LDD971 pKa = 4.57AINGGRR977 pKa = 11.84LSYY980 pKa = 11.27SPFDD984 pKa = 4.68LNDD987 pKa = 3.47DD988 pKa = 3.9TSFDD992 pKa = 3.79DD993 pKa = 3.9GDD995 pKa = 4.17YY996 pKa = 9.96VTVTVEE1002 pKa = 3.79EE1003 pKa = 4.89GEE1005 pKa = 4.31GEE1007 pKa = 4.4EE1008 pKa = 4.11IQVPASGKK1016 pKa = 9.79RR1017 pKa = 11.84LDD1019 pKa = 3.67VGGADD1024 pKa = 3.68MPGVMSGEE1032 pKa = 3.95GSEE1035 pKa = 4.35FKK1037 pKa = 10.95YY1038 pKa = 10.85ISGDD1042 pKa = 3.37DD1043 pKa = 3.48GLQVVRR1049 pKa = 11.84EE1050 pKa = 4.17NPGPEE1055 pKa = 4.01SMGRR1059 pKa = 11.84MSWEE1063 pKa = 3.69QLQQ1066 pKa = 3.47
MM1 pKa = 7.52NRR3 pKa = 11.84FLSFALAVMTALIPAYY19 pKa = 9.82IMAEE23 pKa = 4.02PLDD26 pKa = 4.39LADD29 pKa = 5.36APLATATSAEE39 pKa = 4.41PNVMLLIDD47 pKa = 4.11TSGSMDD53 pKa = 3.78NIIWAEE59 pKa = 4.19GYY61 pKa = 10.03DD62 pKa = 3.84PSVTYY67 pKa = 10.29PGGFDD72 pKa = 4.39ANDD75 pKa = 3.91GNIHH79 pKa = 7.16LYY81 pKa = 8.11ATKK84 pKa = 10.82DD85 pKa = 3.4FGNCGNNGEE94 pKa = 4.25WVEE97 pKa = 4.6DD98 pKa = 3.34QGGWKK103 pKa = 9.74DD104 pKa = 3.62LCLPDD109 pKa = 4.8PVGGDD114 pKa = 3.17ATRR117 pKa = 11.84YY118 pKa = 6.87TGNYY122 pKa = 9.36LNYY125 pKa = 10.17LFYY128 pKa = 10.3IYY130 pKa = 10.58TEE132 pKa = 4.95DD133 pKa = 4.95DD134 pKa = 3.21TDD136 pKa = 3.99LTDD139 pKa = 3.52GTIPTEE145 pKa = 3.82TRR147 pKa = 11.84MQVAKK152 pKa = 10.16EE153 pKa = 3.97VASQLVEE160 pKa = 3.95NTPGMRR166 pKa = 11.84FGVSQFYY173 pKa = 10.75GPTWDD178 pKa = 3.37DD179 pKa = 3.56HH180 pKa = 6.38RR181 pKa = 11.84WGEE184 pKa = 4.09GAEE187 pKa = 4.21VVAACGSSVSDD198 pKa = 3.78LTSEE202 pKa = 4.06IAGFTASTNTPLAEE216 pKa = 4.03ALYY219 pKa = 10.32EE220 pKa = 3.59ITRR223 pKa = 11.84YY224 pKa = 8.95YY225 pKa = 10.91RR226 pKa = 11.84GMDD229 pKa = 3.13AYY231 pKa = 9.62YY232 pKa = 10.53TNDD235 pKa = 3.96DD236 pKa = 3.76FASPIQYY243 pKa = 9.84RR244 pKa = 11.84CQKK247 pKa = 9.8NFTVVISDD255 pKa = 4.11GLPTYY260 pKa = 7.93DD261 pKa = 3.94TNIPGNDD268 pKa = 3.51PAEE271 pKa = 4.4SSGRR275 pKa = 11.84ALPDD279 pKa = 3.2WDD281 pKa = 4.32GLSPDD286 pKa = 4.71TDD288 pKa = 3.76PADD291 pKa = 3.85YY292 pKa = 10.56PDD294 pKa = 4.35FPQYY298 pKa = 11.31SDD300 pKa = 3.83GFQPDD305 pKa = 4.95GYY307 pKa = 9.23PSEE310 pKa = 4.5EE311 pKa = 4.75GYY313 pKa = 10.92SLYY316 pKa = 10.89LDD318 pKa = 5.87DD319 pKa = 4.46IAKK322 pKa = 10.25FGWDD326 pKa = 2.73IDD328 pKa = 3.84LRR330 pKa = 11.84TGGTDD335 pKa = 3.34DD336 pKa = 5.15AGMSFDD342 pKa = 5.59DD343 pKa = 5.17PAFEE347 pKa = 4.0QQNMGTYY354 pKa = 9.99TIGFSTANQMLEE366 pKa = 3.78DD367 pKa = 3.72AAEE370 pKa = 4.32YY371 pKa = 10.79GHH373 pKa = 7.04GEE375 pKa = 4.33YY376 pKa = 9.57LTAANAAEE384 pKa = 4.61LSAALQAAISDD395 pKa = 3.48IAARR399 pKa = 11.84SSSSASATSNTGYY412 pKa = 9.81IASGSRR418 pKa = 11.84VFQARR423 pKa = 11.84YY424 pKa = 9.91NSSNWTGQLLAFAIEE439 pKa = 4.23SDD441 pKa = 3.94VDD443 pKa = 3.86SEE445 pKa = 5.77DD446 pKa = 3.57YY447 pKa = 11.46GFLQRR452 pKa = 11.84NGPVTEE458 pKa = 4.1GAVWDD463 pKa = 3.95AAEE466 pKa = 5.06EE467 pKa = 4.11IPAAGSRR474 pKa = 11.84TIMTYY479 pKa = 10.69NGGGVPFAWTDD490 pKa = 3.31LSTAQQSALGSEE502 pKa = 4.75DD503 pKa = 3.19VLSYY507 pKa = 11.22LRR509 pKa = 11.84GDD511 pKa = 3.46QSQEE515 pKa = 3.72VSSGGSFRR523 pKa = 11.84NRR525 pKa = 11.84DD526 pKa = 3.58ALLGDD531 pKa = 4.06IVNSNPVFMGAPPFNYY547 pKa = 9.52PDD549 pKa = 3.84DD550 pKa = 4.26WNGDD554 pKa = 3.45TTEE557 pKa = 5.02PEE559 pKa = 4.01DD560 pKa = 3.82ASPYY564 pKa = 10.79SSFAVNQNDD573 pKa = 4.5RR574 pKa = 11.84TPMIYY579 pKa = 10.3VGANDD584 pKa = 3.79GMLHH588 pKa = 6.42GFDD591 pKa = 4.66AEE593 pKa = 4.38TGEE596 pKa = 4.08EE597 pKa = 3.79RR598 pKa = 11.84MAYY601 pKa = 9.96VPGSVFEE608 pKa = 4.57NLDD611 pKa = 3.57EE612 pKa = 5.82LVDD615 pKa = 3.13TDD617 pKa = 3.68YY618 pKa = 11.72AHH620 pKa = 7.85RR621 pKa = 11.84YY622 pKa = 8.26FVDD625 pKa = 4.88GSPTIADD632 pKa = 3.75AYY634 pKa = 11.09FDD636 pKa = 4.61GSWHH640 pKa = 5.61TVLLGGLNKK649 pKa = 10.28GGQGIYY655 pKa = 10.77ALDD658 pKa = 3.48VTTATFSEE666 pKa = 4.89GNADD670 pKa = 4.04STVLWEE676 pKa = 4.38FTDD679 pKa = 4.63ADD681 pKa = 4.42DD682 pKa = 4.67VDD684 pKa = 4.04LGYY687 pKa = 9.28TFSQPNIVRR696 pKa = 11.84LHH698 pKa = 5.72NGQWAAIFGNGYY710 pKa = 10.24NNTRR714 pKa = 11.84ADD716 pKa = 3.7GDD718 pKa = 3.67ASTTGHH724 pKa = 6.12AVLYY728 pKa = 9.9IVSIEE733 pKa = 4.21DD734 pKa = 3.12GSLIRR739 pKa = 11.84KK740 pKa = 8.99IDD742 pKa = 3.57TGAGTATTPNGLATVSPVDD761 pKa = 3.19VDD763 pKa = 2.83GDD765 pKa = 4.35YY766 pKa = 11.1IVDD769 pKa = 3.54RR770 pKa = 11.84VYY772 pKa = 11.24AGDD775 pKa = 3.58LQGNLWRR782 pKa = 11.84FDD784 pKa = 4.29LDD786 pKa = 4.97DD787 pKa = 4.83GNPNQWGIPFSQGQTHH803 pKa = 6.41EE804 pKa = 4.61PLFSACSGTPCDD816 pKa = 5.01SNRR819 pKa = 11.84QPITVRR825 pKa = 11.84PEE827 pKa = 3.28VGRR830 pKa = 11.84APNKK834 pKa = 9.46PGAVMLYY841 pKa = 10.04FGTGKK846 pKa = 10.44YY847 pKa = 10.33LEE849 pKa = 4.75NGDD852 pKa = 4.25GNVADD857 pKa = 4.41TPQMQRR863 pKa = 11.84FYY865 pKa = 11.59GVLDD869 pKa = 3.64EE870 pKa = 6.03DD871 pKa = 4.72LPANQFDD878 pKa = 4.14TLVPGDD884 pKa = 4.42LLEE887 pKa = 4.17QEE889 pKa = 4.68ILVEE893 pKa = 4.13SSFNVEE899 pKa = 4.37GNSHH903 pKa = 5.63SLRR906 pKa = 11.84VTSDD910 pKa = 3.27YY911 pKa = 11.52TLGSEE916 pKa = 4.68DD917 pKa = 3.68KK918 pKa = 10.83GWYY921 pKa = 8.96IDD923 pKa = 4.05LAPPGTGSVGEE934 pKa = 4.25IAVTAPLLRR943 pKa = 11.84AGRR946 pKa = 11.84VFFTTLIPGEE956 pKa = 4.88DD957 pKa = 3.53PCQPGGQGWLMEE969 pKa = 4.68LDD971 pKa = 4.57AINGGRR977 pKa = 11.84LSYY980 pKa = 11.27SPFDD984 pKa = 4.68LNDD987 pKa = 3.47DD988 pKa = 3.9TSFDD992 pKa = 3.79DD993 pKa = 3.9GDD995 pKa = 4.17YY996 pKa = 9.96VTVTVEE1002 pKa = 3.79EE1003 pKa = 4.89GEE1005 pKa = 4.31GEE1007 pKa = 4.4EE1008 pKa = 4.11IQVPASGKK1016 pKa = 9.79RR1017 pKa = 11.84LDD1019 pKa = 3.67VGGADD1024 pKa = 3.68MPGVMSGEE1032 pKa = 3.95GSEE1035 pKa = 4.35FKK1037 pKa = 10.95YY1038 pKa = 10.85ISGDD1042 pKa = 3.37DD1043 pKa = 3.48GLQVVRR1049 pKa = 11.84EE1050 pKa = 4.17NPGPEE1055 pKa = 4.01SMGRR1059 pKa = 11.84MSWEE1063 pKa = 3.69QLQQ1066 pKa = 3.47
Molecular weight: 114.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I4R1B2|A0A1I4R1B2_ECTMO Delta-aminolevulinic acid dehydratase OS=Ectothiorhodospira mobilis OX=195064 GN=SAMN05421721_10669 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLHH11 pKa = 6.3RR12 pKa = 11.84KK13 pKa = 6.84RR14 pKa = 11.84THH16 pKa = 5.74GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84GGRR28 pKa = 11.84KK29 pKa = 8.98VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.28GRR39 pKa = 11.84ARR41 pKa = 11.84LCPP44 pKa = 3.71
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLHH11 pKa = 6.3RR12 pKa = 11.84KK13 pKa = 6.84RR14 pKa = 11.84THH16 pKa = 5.74GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84GGRR28 pKa = 11.84KK29 pKa = 8.98VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.28GRR39 pKa = 11.84ARR41 pKa = 11.84LCPP44 pKa = 3.71
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
789774 |
29 |
1816 |
327.3 |
36.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.315 ± 0.063 | 0.936 ± 0.018 |
5.763 ± 0.04 | 6.769 ± 0.053 |
3.173 ± 0.029 | 8.788 ± 0.05 |
2.637 ± 0.027 | 4.087 ± 0.035 |
2.171 ± 0.036 | 11.46 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.426 ± 0.022 | 2.081 ± 0.025 |
5.668 ± 0.041 | 3.869 ± 0.032 |
8.925 ± 0.048 | 4.288 ± 0.025 |
4.736 ± 0.03 | 7.349 ± 0.044 |
1.34 ± 0.02 | 2.221 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
