
Dragonfly-associated circular virus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus draga1; Dragonfly associated gemycircularvirus 1
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
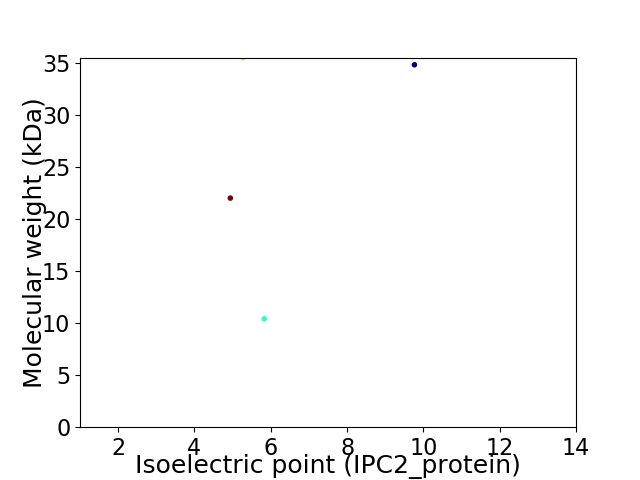
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K0A2R4|K0A2R4_9VIRU Replication-associated protein OS=Dragonfly-associated circular virus 2 OX=1234886 PE=3 SV=1
MM1 pKa = 7.81PSFVCNARR9 pKa = 11.84YY10 pKa = 9.04FLVTYY15 pKa = 7.11PQCGNLCGFSVVEE28 pKa = 4.33RR29 pKa = 11.84FSSLGAEE36 pKa = 4.2CIVGRR41 pKa = 11.84EE42 pKa = 3.95HH43 pKa = 8.11HH44 pKa = 6.88EE45 pKa = 4.43DD46 pKa = 3.48GGLHH50 pKa = 5.8LHH52 pKa = 6.83CFADD56 pKa = 4.75FGRR59 pKa = 11.84KK60 pKa = 8.15FRR62 pKa = 11.84SRR64 pKa = 11.84KK65 pKa = 7.67TDD67 pKa = 2.69IFDD70 pKa = 4.44VDD72 pKa = 3.66GCHH75 pKa = 7.11PNIQPSTGTPEE86 pKa = 3.53KK87 pKa = 10.7GYY89 pKa = 10.88DD90 pKa = 3.47YY91 pKa = 10.69AIKK94 pKa = 10.87DD95 pKa = 3.59GDD97 pKa = 4.01VVAGGLEE104 pKa = 3.96RR105 pKa = 11.84PIGKK109 pKa = 9.14SRR111 pKa = 11.84AGARR115 pKa = 11.84TTFDD119 pKa = 2.52KK120 pKa = 10.51WAEE123 pKa = 3.75ITGAEE128 pKa = 4.61DD129 pKa = 5.92RR130 pKa = 11.84DD131 pKa = 4.54TFWQLCHH138 pKa = 6.67EE139 pKa = 5.52LDD141 pKa = 4.23PKK143 pKa = 10.38ATACSFGQLSKK154 pKa = 10.58YY155 pKa = 10.59ADD157 pKa = 2.97WRR159 pKa = 11.84FAEE162 pKa = 4.65VPAEE166 pKa = 3.97YY167 pKa = 9.83EE168 pKa = 4.28SPGGLEE174 pKa = 4.46FIGGDD179 pKa = 3.16VDD181 pKa = 6.0GRR183 pKa = 11.84DD184 pKa = 3.69DD185 pKa = 3.57WVQSAGIGLGQSFVGGG201 pKa = 3.64
MM1 pKa = 7.81PSFVCNARR9 pKa = 11.84YY10 pKa = 9.04FLVTYY15 pKa = 7.11PQCGNLCGFSVVEE28 pKa = 4.33RR29 pKa = 11.84FSSLGAEE36 pKa = 4.2CIVGRR41 pKa = 11.84EE42 pKa = 3.95HH43 pKa = 8.11HH44 pKa = 6.88EE45 pKa = 4.43DD46 pKa = 3.48GGLHH50 pKa = 5.8LHH52 pKa = 6.83CFADD56 pKa = 4.75FGRR59 pKa = 11.84KK60 pKa = 8.15FRR62 pKa = 11.84SRR64 pKa = 11.84KK65 pKa = 7.67TDD67 pKa = 2.69IFDD70 pKa = 4.44VDD72 pKa = 3.66GCHH75 pKa = 7.11PNIQPSTGTPEE86 pKa = 3.53KK87 pKa = 10.7GYY89 pKa = 10.88DD90 pKa = 3.47YY91 pKa = 10.69AIKK94 pKa = 10.87DD95 pKa = 3.59GDD97 pKa = 4.01VVAGGLEE104 pKa = 3.96RR105 pKa = 11.84PIGKK109 pKa = 9.14SRR111 pKa = 11.84AGARR115 pKa = 11.84TTFDD119 pKa = 2.52KK120 pKa = 10.51WAEE123 pKa = 3.75ITGAEE128 pKa = 4.61DD129 pKa = 5.92RR130 pKa = 11.84DD131 pKa = 4.54TFWQLCHH138 pKa = 6.67EE139 pKa = 5.52LDD141 pKa = 4.23PKK143 pKa = 10.38ATACSFGQLSKK154 pKa = 10.58YY155 pKa = 10.59ADD157 pKa = 2.97WRR159 pKa = 11.84FAEE162 pKa = 4.65VPAEE166 pKa = 3.97YY167 pKa = 9.83EE168 pKa = 4.28SPGGLEE174 pKa = 4.46FIGGDD179 pKa = 3.16VDD181 pKa = 6.0GRR183 pKa = 11.84DD184 pKa = 3.69DD185 pKa = 3.57WVQSAGIGLGQSFVGGG201 pKa = 3.64
Molecular weight: 22.0 kDa
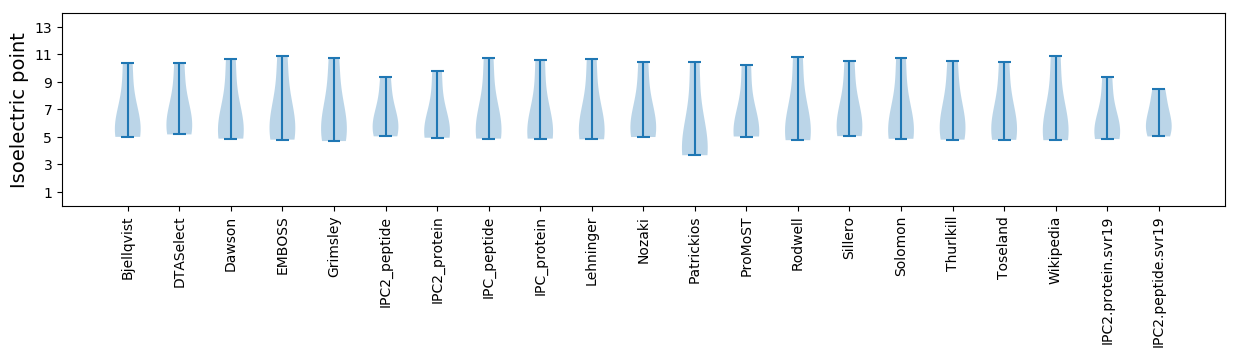
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K0A172|K0A172_9VIRU Replication-associated protein OS=Dragonfly-associated circular virus 2 OX=1234886 PE=4 SV=1
MM1 pKa = 7.74AYY3 pKa = 10.14ARR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84SGSYY11 pKa = 9.12RR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84QTKK18 pKa = 9.77RR19 pKa = 11.84RR20 pKa = 11.84GTGYY24 pKa = 9.98RR25 pKa = 11.84RR26 pKa = 11.84SSAKK30 pKa = 7.15TRR32 pKa = 11.84RR33 pKa = 11.84YY34 pKa = 10.02RR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 8.45RR39 pKa = 11.84TSRR42 pKa = 11.84PMSKK46 pKa = 9.64RR47 pKa = 11.84RR48 pKa = 11.84MLNVSSRR55 pKa = 11.84KK56 pKa = 9.33KK57 pKa = 10.14RR58 pKa = 11.84NGMLSWSNTASTGASQTVASGPAYY82 pKa = 11.06VNGTGTGFFLWNATANDD99 pKa = 4.14LNSGNGAYY107 pKa = 8.32NTIAQEE113 pKa = 4.23SQRR116 pKa = 11.84TSTTCYY122 pKa = 8.95MRR124 pKa = 11.84GLSEE128 pKa = 5.06HH129 pKa = 6.59IRR131 pKa = 11.84IQTSSGLPWFWRR143 pKa = 11.84RR144 pKa = 11.84ICFTFRR150 pKa = 11.84GSFPAASDD158 pKa = 3.95SPTQTPRR165 pKa = 11.84NFQDD169 pKa = 3.09TSNGIVRR176 pKa = 11.84LWFNTDD182 pKa = 2.87VNNTPNLRR190 pKa = 11.84NQVDD194 pKa = 3.9GVIFKK199 pKa = 10.19GAKK202 pKa = 9.04GVDD205 pKa = 3.05WNDD208 pKa = 4.11FIVAPVDD215 pKa = 3.41TRR217 pKa = 11.84RR218 pKa = 11.84VDD220 pKa = 3.72LKK222 pKa = 10.86YY223 pKa = 11.03DD224 pKa = 3.0RR225 pKa = 11.84TMCIRR230 pKa = 11.84SGNAAGTVKK239 pKa = 10.09EE240 pKa = 4.1AKK242 pKa = 9.2LWHH245 pKa = 6.12PMNRR249 pKa = 11.84NLVYY253 pKa = 10.84DD254 pKa = 4.54DD255 pKa = 5.66DD256 pKa = 4.69EE257 pKa = 6.97SGDD260 pKa = 3.88QEE262 pKa = 4.15ATSYY266 pKa = 11.48LSVEE270 pKa = 4.7SKK272 pKa = 11.19AGMGDD277 pKa = 4.12YY278 pKa = 10.84YY279 pKa = 11.67VLDD282 pKa = 4.3LLQAGSGGSNSDD294 pKa = 4.45QILLQANSTLYY305 pKa = 9.39WHH307 pKa = 7.05EE308 pKa = 4.08KK309 pKa = 9.32
MM1 pKa = 7.74AYY3 pKa = 10.14ARR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84SGSYY11 pKa = 9.12RR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84QTKK18 pKa = 9.77RR19 pKa = 11.84RR20 pKa = 11.84GTGYY24 pKa = 9.98RR25 pKa = 11.84RR26 pKa = 11.84SSAKK30 pKa = 7.15TRR32 pKa = 11.84RR33 pKa = 11.84YY34 pKa = 10.02RR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 8.45RR39 pKa = 11.84TSRR42 pKa = 11.84PMSKK46 pKa = 9.64RR47 pKa = 11.84RR48 pKa = 11.84MLNVSSRR55 pKa = 11.84KK56 pKa = 9.33KK57 pKa = 10.14RR58 pKa = 11.84NGMLSWSNTASTGASQTVASGPAYY82 pKa = 11.06VNGTGTGFFLWNATANDD99 pKa = 4.14LNSGNGAYY107 pKa = 8.32NTIAQEE113 pKa = 4.23SQRR116 pKa = 11.84TSTTCYY122 pKa = 8.95MRR124 pKa = 11.84GLSEE128 pKa = 5.06HH129 pKa = 6.59IRR131 pKa = 11.84IQTSSGLPWFWRR143 pKa = 11.84RR144 pKa = 11.84ICFTFRR150 pKa = 11.84GSFPAASDD158 pKa = 3.95SPTQTPRR165 pKa = 11.84NFQDD169 pKa = 3.09TSNGIVRR176 pKa = 11.84LWFNTDD182 pKa = 2.87VNNTPNLRR190 pKa = 11.84NQVDD194 pKa = 3.9GVIFKK199 pKa = 10.19GAKK202 pKa = 9.04GVDD205 pKa = 3.05WNDD208 pKa = 4.11FIVAPVDD215 pKa = 3.41TRR217 pKa = 11.84RR218 pKa = 11.84VDD220 pKa = 3.72LKK222 pKa = 10.86YY223 pKa = 11.03DD224 pKa = 3.0RR225 pKa = 11.84TMCIRR230 pKa = 11.84SGNAAGTVKK239 pKa = 10.09EE240 pKa = 4.1AKK242 pKa = 9.2LWHH245 pKa = 6.12PMNRR249 pKa = 11.84NLVYY253 pKa = 10.84DD254 pKa = 4.54DD255 pKa = 5.66DD256 pKa = 4.69EE257 pKa = 6.97SGDD260 pKa = 3.88QEE262 pKa = 4.15ATSYY266 pKa = 11.48LSVEE270 pKa = 4.7SKK272 pKa = 11.19AGMGDD277 pKa = 4.12YY278 pKa = 10.84YY279 pKa = 11.67VLDD282 pKa = 4.3LLQAGSGGSNSDD294 pKa = 4.45QILLQANSTLYY305 pKa = 9.39WHH307 pKa = 7.05EE308 pKa = 4.08KK309 pKa = 9.32
Molecular weight: 34.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
920 |
91 |
319 |
230.0 |
25.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.609 ± 0.138 | 2.174 ± 0.614 |
7.065 ± 0.565 | 5.0 ± 1.018 |
5.326 ± 0.74 | 9.891 ± 1.0 |
2.174 ± 0.451 | 4.457 ± 0.67 |
4.565 ± 0.271 | 5.87 ± 0.162 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.522 ± 0.437 | 4.022 ± 1.175 |
4.348 ± 0.53 | 2.717 ± 0.48 |
7.717 ± 1.161 | 8.043 ± 0.853 |
5.543 ± 1.133 | 5.87 ± 0.393 |
2.717 ± 0.241 | 3.37 ± 0.293 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
