
Silicibacter phage DSS3-P1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
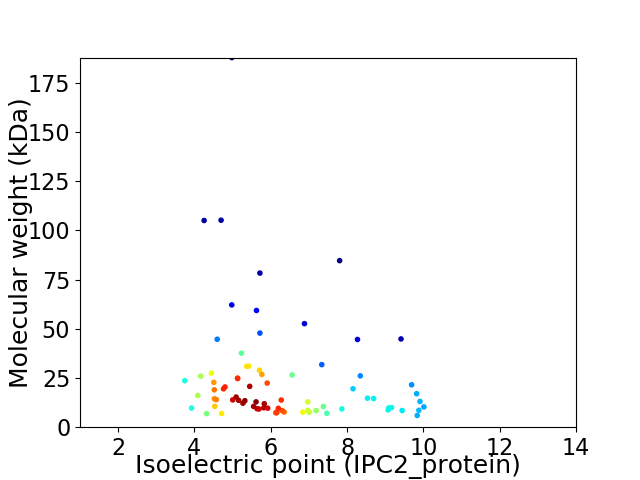
Virtual 2D-PAGE plot for 78 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G8DCV7|G8DCV7_9CAUD DNA methyltransferase OS=Silicibacter phage DSS3-P1 OX=754060 GN=SDSG_00072 PE=3 SV=1
MM1 pKa = 7.46SNTYY5 pKa = 8.72TVAVVEE11 pKa = 4.9GSTPLPPCYY20 pKa = 9.19PDD22 pKa = 3.63PLLYY26 pKa = 10.62VVTVSDD32 pKa = 3.7PHH34 pKa = 8.21DD35 pKa = 4.06SEE37 pKa = 6.28SILDD41 pKa = 3.7QVAEE45 pKa = 4.07ARR47 pKa = 11.84ALEE50 pKa = 4.07LLGEE54 pKa = 4.49PEE56 pKa = 5.59DD57 pKa = 4.07YY58 pKa = 10.73TPQTFDD64 pKa = 3.39ALRR67 pKa = 11.84AGLRR71 pKa = 11.84LLFVLAGDD79 pKa = 4.45VYY81 pKa = 10.73PVADD85 pKa = 4.78FRR87 pKa = 11.84QSS89 pKa = 2.73
MM1 pKa = 7.46SNTYY5 pKa = 8.72TVAVVEE11 pKa = 4.9GSTPLPPCYY20 pKa = 9.19PDD22 pKa = 3.63PLLYY26 pKa = 10.62VVTVSDD32 pKa = 3.7PHH34 pKa = 8.21DD35 pKa = 4.06SEE37 pKa = 6.28SILDD41 pKa = 3.7QVAEE45 pKa = 4.07ARR47 pKa = 11.84ALEE50 pKa = 4.07LLGEE54 pKa = 4.49PEE56 pKa = 5.59DD57 pKa = 4.07YY58 pKa = 10.73TPQTFDD64 pKa = 3.39ALRR67 pKa = 11.84AGLRR71 pKa = 11.84LLFVLAGDD79 pKa = 4.45VYY81 pKa = 10.73PVADD85 pKa = 4.78FRR87 pKa = 11.84QSS89 pKa = 2.73
Molecular weight: 9.73 kDa
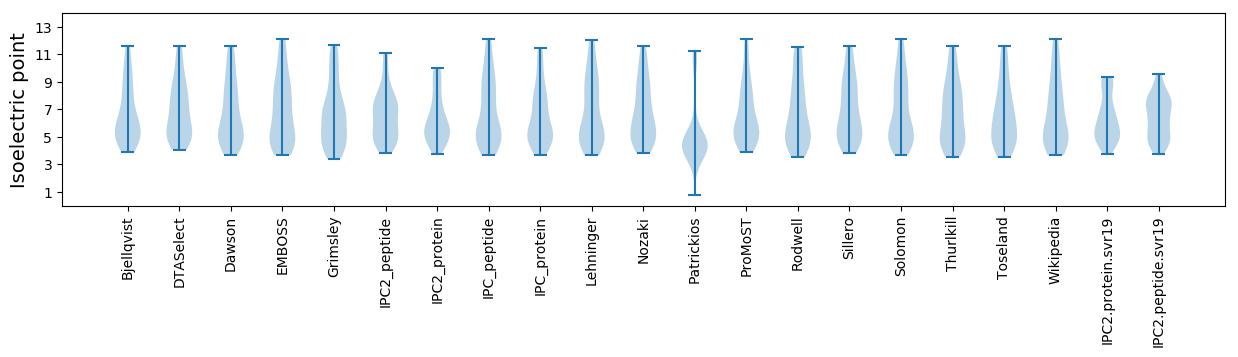
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8DCQ9|G8DCQ9_9CAUD Uncharacterized protein OS=Silicibacter phage DSS3-P1 OX=754060 GN=SDSG_00023 PE=4 SV=1
MM1 pKa = 7.53NLPDD5 pKa = 4.36TNPKK9 pKa = 8.7TRR11 pKa = 11.84FGMAKK16 pKa = 9.21PGIYY20 pKa = 10.11AIPPTALLHH29 pKa = 6.65CGAAMHH35 pKa = 6.51NGCEE39 pKa = 4.67KK40 pKa = 11.02YY41 pKa = 10.87GLTNWRR47 pKa = 11.84EE48 pKa = 4.25HH49 pKa = 5.82EE50 pKa = 4.43VSASVYY56 pKa = 8.31YY57 pKa = 10.57NAAFRR62 pKa = 11.84HH63 pKa = 5.37LAAWWDD69 pKa = 3.84GEE71 pKa = 4.59RR72 pKa = 11.84EE73 pKa = 4.22AEE75 pKa = 4.25DD76 pKa = 3.6SGVHH80 pKa = 5.84HH81 pKa = 7.46LGHH84 pKa = 6.81VMACCAVLLDD94 pKa = 4.27AEE96 pKa = 4.68HH97 pKa = 6.37MGKK100 pKa = 9.71LRR102 pKa = 11.84DD103 pKa = 4.35DD104 pKa = 3.86RR105 pKa = 11.84PAIPGPFSFAVAEE118 pKa = 4.2MTKK121 pKa = 10.08PQAEE125 pKa = 4.32EE126 pKa = 5.15GKK128 pKa = 7.95QTWGGFCQDD137 pKa = 2.83IAASAEE143 pKa = 3.85AAFAKK148 pKa = 10.59HH149 pKa = 5.81EE150 pKa = 4.34ADD152 pKa = 3.57EE153 pKa = 4.12QLEE156 pKa = 4.13ADD158 pKa = 4.33CEE160 pKa = 4.29EE161 pKa = 3.86IALRR165 pKa = 11.84VLSTGSLDD173 pKa = 3.5DD174 pKa = 3.87AKK176 pKa = 11.19AVIRR180 pKa = 11.84AYY182 pKa = 9.74LTGGGTTRR190 pKa = 11.84KK191 pKa = 9.64ISGRR195 pKa = 11.84LLATPSIGKK204 pKa = 8.45MSRR207 pKa = 11.84PRR209 pKa = 11.84STLSRR214 pKa = 11.84RR215 pKa = 11.84PRR217 pKa = 11.84TPVSPLGSTQARR229 pKa = 11.84APEE232 pKa = 3.89PAMTGMLSSRR242 pKa = 11.84SSRR245 pKa = 11.84RR246 pKa = 11.84LPTLNWSRR254 pKa = 11.84ASGRR258 pKa = 11.84SWIKK262 pKa = 9.7TGPDD266 pKa = 3.16GSRR269 pKa = 11.84SLRR272 pKa = 11.84FSATTSVPSGSRR284 pKa = 11.84PTVRR288 pKa = 11.84TTGPGSRR295 pKa = 11.84STSDD299 pKa = 2.82ARR301 pKa = 11.84PRR303 pKa = 11.84DD304 pKa = 3.79STHH307 pKa = 7.1RR308 pKa = 11.84PPSWTMPSAWPVSSSEE324 pKa = 3.98PPDD327 pKa = 2.98RR328 pKa = 11.84CRR330 pKa = 11.84PLTPDD335 pKa = 2.83SPRR338 pKa = 11.84PPRR341 pKa = 11.84IAGCVAPAPSACRR354 pKa = 11.84TRR356 pKa = 11.84SRR358 pKa = 11.84KK359 pKa = 9.13RR360 pKa = 11.84APASSPPRR368 pKa = 11.84GPWRR372 pKa = 11.84TTSGRR377 pKa = 11.84CAWFSAWSRR386 pKa = 11.84RR387 pKa = 11.84ISWDD391 pKa = 3.09RR392 pKa = 11.84KK393 pKa = 7.67SEE395 pKa = 4.14RR396 pKa = 11.84TVSPSPSRR404 pKa = 11.84TRR406 pKa = 11.84WWRR409 pKa = 11.84RR410 pKa = 2.92
MM1 pKa = 7.53NLPDD5 pKa = 4.36TNPKK9 pKa = 8.7TRR11 pKa = 11.84FGMAKK16 pKa = 9.21PGIYY20 pKa = 10.11AIPPTALLHH29 pKa = 6.65CGAAMHH35 pKa = 6.51NGCEE39 pKa = 4.67KK40 pKa = 11.02YY41 pKa = 10.87GLTNWRR47 pKa = 11.84EE48 pKa = 4.25HH49 pKa = 5.82EE50 pKa = 4.43VSASVYY56 pKa = 8.31YY57 pKa = 10.57NAAFRR62 pKa = 11.84HH63 pKa = 5.37LAAWWDD69 pKa = 3.84GEE71 pKa = 4.59RR72 pKa = 11.84EE73 pKa = 4.22AEE75 pKa = 4.25DD76 pKa = 3.6SGVHH80 pKa = 5.84HH81 pKa = 7.46LGHH84 pKa = 6.81VMACCAVLLDD94 pKa = 4.27AEE96 pKa = 4.68HH97 pKa = 6.37MGKK100 pKa = 9.71LRR102 pKa = 11.84DD103 pKa = 4.35DD104 pKa = 3.86RR105 pKa = 11.84PAIPGPFSFAVAEE118 pKa = 4.2MTKK121 pKa = 10.08PQAEE125 pKa = 4.32EE126 pKa = 5.15GKK128 pKa = 7.95QTWGGFCQDD137 pKa = 2.83IAASAEE143 pKa = 3.85AAFAKK148 pKa = 10.59HH149 pKa = 5.81EE150 pKa = 4.34ADD152 pKa = 3.57EE153 pKa = 4.12QLEE156 pKa = 4.13ADD158 pKa = 4.33CEE160 pKa = 4.29EE161 pKa = 3.86IALRR165 pKa = 11.84VLSTGSLDD173 pKa = 3.5DD174 pKa = 3.87AKK176 pKa = 11.19AVIRR180 pKa = 11.84AYY182 pKa = 9.74LTGGGTTRR190 pKa = 11.84KK191 pKa = 9.64ISGRR195 pKa = 11.84LLATPSIGKK204 pKa = 8.45MSRR207 pKa = 11.84PRR209 pKa = 11.84STLSRR214 pKa = 11.84RR215 pKa = 11.84PRR217 pKa = 11.84TPVSPLGSTQARR229 pKa = 11.84APEE232 pKa = 3.89PAMTGMLSSRR242 pKa = 11.84SSRR245 pKa = 11.84RR246 pKa = 11.84LPTLNWSRR254 pKa = 11.84ASGRR258 pKa = 11.84SWIKK262 pKa = 9.7TGPDD266 pKa = 3.16GSRR269 pKa = 11.84SLRR272 pKa = 11.84FSATTSVPSGSRR284 pKa = 11.84PTVRR288 pKa = 11.84TTGPGSRR295 pKa = 11.84STSDD299 pKa = 2.82ARR301 pKa = 11.84PRR303 pKa = 11.84DD304 pKa = 3.79STHH307 pKa = 7.1RR308 pKa = 11.84PPSWTMPSAWPVSSSEE324 pKa = 3.98PPDD327 pKa = 2.98RR328 pKa = 11.84CRR330 pKa = 11.84PLTPDD335 pKa = 2.83SPRR338 pKa = 11.84PPRR341 pKa = 11.84IAGCVAPAPSACRR354 pKa = 11.84TRR356 pKa = 11.84SRR358 pKa = 11.84KK359 pKa = 9.13RR360 pKa = 11.84APASSPPRR368 pKa = 11.84GPWRR372 pKa = 11.84TTSGRR377 pKa = 11.84CAWFSAWSRR386 pKa = 11.84RR387 pKa = 11.84ISWDD391 pKa = 3.09RR392 pKa = 11.84KK393 pKa = 7.67SEE395 pKa = 4.14RR396 pKa = 11.84TVSPSPSRR404 pKa = 11.84TRR406 pKa = 11.84WWRR409 pKa = 11.84RR410 pKa = 2.92
Molecular weight: 44.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
17487 |
54 |
1744 |
224.2 |
24.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.62 ± 0.628 | 0.858 ± 0.11 |
6.496 ± 0.255 | 7.005 ± 0.272 |
3.362 ± 0.162 | 8.12 ± 0.276 |
2.047 ± 0.196 | 4.34 ± 0.177 |
3.923 ± 0.318 | 8.595 ± 0.243 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.327 ± 0.162 | 3.162 ± 0.165 |
5.227 ± 0.431 | 3.38 ± 0.288 |
7.566 ± 0.32 | 5.267 ± 0.228 |
5.999 ± 0.283 | 6.473 ± 0.194 |
1.698 ± 0.185 | 2.533 ± 0.174 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
