
Gracilimonas mengyeensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Balneolaeota; Balneolia; Balneolales; Balneolaceae; Gracilimonas
Average proteome isoelectric point is 5.7
Get precalculated fractions of proteins
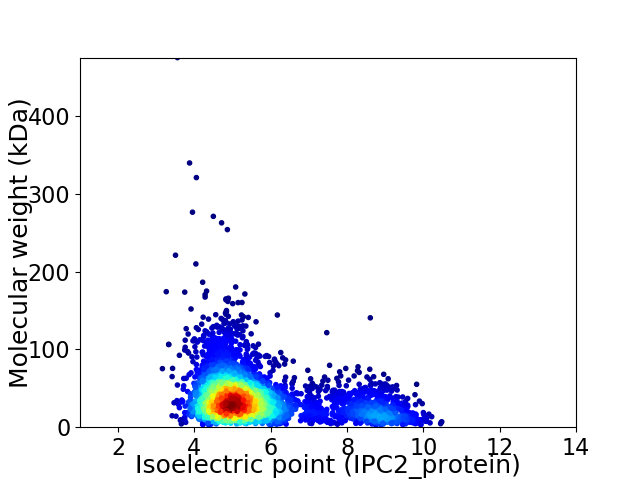
Virtual 2D-PAGE plot for 3810 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
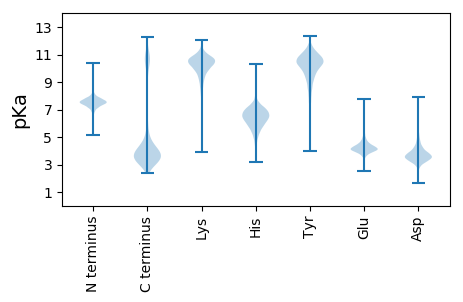
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A521C3D4|A0A521C3D4_9BACT Uncharacterized protein OS=Gracilimonas mengyeensis OX=1302730 GN=SAMN06265219_104120 PE=4 SV=1
MM1 pKa = 7.81GIFPYY6 pKa = 10.55AQAQEE11 pKa = 4.36VVTTFDD17 pKa = 4.39LNGADD22 pKa = 3.11GTLYY26 pKa = 10.77NDD28 pKa = 3.91EE29 pKa = 4.29NRR31 pKa = 11.84GADD34 pKa = 3.62FPGGDD39 pKa = 3.42RR40 pKa = 11.84SSLEE44 pKa = 3.14IRR46 pKa = 11.84YY47 pKa = 8.55WDD49 pKa = 3.45EE50 pKa = 3.6VRR52 pKa = 11.84FRR54 pKa = 11.84ASMVRR59 pKa = 11.84FDD61 pKa = 3.93VNAVNSSPDD70 pKa = 3.41GAVLGLYY77 pKa = 6.87PTSAGSMGADD87 pKa = 2.53SLTFTIYY94 pKa = 11.2GLTNEE99 pKa = 5.67SEE101 pKa = 4.7DD102 pKa = 3.49NWDD105 pKa = 3.74EE106 pKa = 4.37GSFSYY111 pKa = 11.3NEE113 pKa = 3.94APGFEE118 pKa = 4.04AAANGTYY125 pKa = 10.29AINEE129 pKa = 4.35DD130 pKa = 4.4LDD132 pKa = 3.9SLTTITVYY140 pKa = 11.35NNLTDD145 pKa = 3.44TYY147 pKa = 10.66LYY149 pKa = 10.47SEE151 pKa = 5.19PSAEE155 pKa = 4.21LDD157 pKa = 3.42AFINSDD163 pKa = 3.67TNGLVTFAIVKK174 pKa = 8.7TYY176 pKa = 8.63STEE179 pKa = 3.77SGSYY183 pKa = 9.91VISPKK188 pKa = 10.36EE189 pKa = 3.86AGEE192 pKa = 4.28SVAPKK197 pKa = 10.26LVYY200 pKa = 10.12QSEE203 pKa = 4.32NGEE206 pKa = 3.88PVLPNTEE213 pKa = 3.85PSITLDD219 pKa = 3.53SPTDD223 pKa = 3.59GASYY227 pKa = 10.59EE228 pKa = 4.52SNNTLTASASASDD241 pKa = 3.82SDD243 pKa = 4.64GEE245 pKa = 4.11IASVSFYY252 pKa = 11.17LDD254 pKa = 3.19DD255 pKa = 4.0TEE257 pKa = 5.04IGSVNASPFEE267 pKa = 4.0YY268 pKa = 10.47EE269 pKa = 4.31IDD271 pKa = 3.41LSMYY275 pKa = 11.01ALGSHH280 pKa = 6.47EE281 pKa = 4.84FYY283 pKa = 10.85AIAEE287 pKa = 4.53DD288 pKa = 3.58NDD290 pKa = 4.01GAVASSDD297 pKa = 2.95TVMFEE302 pKa = 3.34ITEE305 pKa = 4.74AIGGSDD311 pKa = 3.57AEE313 pKa = 4.32MVTTFEE319 pKa = 5.57LNGADD324 pKa = 3.21AAIYY328 pKa = 10.86NDD330 pKa = 4.0DD331 pKa = 4.2NRR333 pKa = 11.84GADD336 pKa = 3.46YY337 pKa = 10.84TGGNRR342 pKa = 11.84SDD344 pKa = 3.97LEE346 pKa = 3.55IRR348 pKa = 11.84YY349 pKa = 8.63WEE351 pKa = 4.1EE352 pKa = 3.3VRR354 pKa = 11.84FRR356 pKa = 11.84ASILRR361 pKa = 11.84FDD363 pKa = 4.01VSNVSSTPNGAVIGFNPTYY382 pKa = 9.47AGKK385 pKa = 9.24MGAEE389 pKa = 4.06SLTYY393 pKa = 9.73TIYY396 pKa = 11.33GMTDD400 pKa = 3.17EE401 pKa = 5.82SQDD404 pKa = 3.28NWDD407 pKa = 4.08EE408 pKa = 4.17GSVNYY413 pKa = 10.46SSAPGFEE420 pKa = 4.36SAANGTYY427 pKa = 11.19ALTEE431 pKa = 4.14NMDD434 pKa = 3.96SLTTITVYY442 pKa = 11.15ADD444 pKa = 3.02STGAFLYY451 pKa = 10.48SDD453 pKa = 4.62PSTEE457 pKa = 4.2LDD459 pKa = 3.22AFINNDD465 pKa = 3.25SNGLVTFAIIKK476 pKa = 8.02TISTNSDD483 pKa = 3.64YY484 pKa = 11.66YY485 pKa = 11.89AMNSKK490 pKa = 10.02EE491 pKa = 4.18AGVSVAPRR499 pKa = 11.84LVYY502 pKa = 10.33QSEE505 pKa = 4.15NGEE508 pKa = 3.83PVLPNFEE515 pKa = 4.23PSITLNSPEE524 pKa = 4.31EE525 pKa = 4.11GMVYY529 pKa = 10.43QSDD532 pKa = 3.83STLTARR538 pKa = 11.84ATAIDD543 pKa = 3.86SDD545 pKa = 4.59GEE547 pKa = 4.04IASVSFYY554 pKa = 10.55MDD556 pKa = 3.52GTAVANVTSSPYY568 pKa = 10.15EE569 pKa = 4.01SEE571 pKa = 4.32IDD573 pKa = 3.36LSMYY577 pKa = 11.03ASGSYY582 pKa = 9.67EE583 pKa = 3.93FYY585 pKa = 10.83AIAEE589 pKa = 4.59DD590 pKa = 4.87DD591 pKa = 4.46DD592 pKa = 4.59GATASSDD599 pKa = 3.25TVTIDD604 pKa = 2.94IVGGSGAGSSEE615 pKa = 3.94MVTTFDD621 pKa = 4.66SNGADD626 pKa = 2.97VTLFNDD632 pKa = 3.55EE633 pKa = 3.94NRR635 pKa = 11.84AGSFTGGDD643 pKa = 3.23RR644 pKa = 11.84TDD646 pKa = 3.56LEE648 pKa = 4.06VRR650 pKa = 11.84YY651 pKa = 9.19WEE653 pKa = 4.03AVRR656 pKa = 11.84FRR658 pKa = 11.84ASMLRR663 pKa = 11.84FDD665 pKa = 4.29VSSVSAAPDD674 pKa = 3.46GAVIGINPTYY684 pKa = 10.4AEE686 pKa = 4.07NMSDD690 pKa = 3.29EE691 pKa = 4.5TLSFTIFGLTNEE703 pKa = 5.5AEE705 pKa = 4.42DD706 pKa = 3.78EE707 pKa = 3.99WDD709 pKa = 3.72EE710 pKa = 4.8GSISYY715 pKa = 10.81NSAPGFEE722 pKa = 4.45SAINGSYY729 pKa = 10.59AINDD733 pKa = 4.15DD734 pKa = 4.36LDD736 pKa = 3.82SLTTITVYY744 pKa = 11.14ADD746 pKa = 3.13STGTFLYY753 pKa = 10.57SEE755 pKa = 4.96PSDD758 pKa = 3.75KK759 pKa = 11.07LDD761 pKa = 3.72AFIRR765 pKa = 11.84NDD767 pKa = 3.23NNGLVTFAIIRR778 pKa = 11.84TYY780 pKa = 11.43SNNSDD785 pKa = 3.18IYY787 pKa = 11.42AMSSKK792 pKa = 10.42EE793 pKa = 3.87AGEE796 pKa = 4.01LVAPRR801 pKa = 11.84LVYY804 pKa = 10.38KK805 pKa = 10.5SEE807 pKa = 4.04NGVPVIPNLEE817 pKa = 4.1PNITLSRR824 pKa = 11.84PADD827 pKa = 3.57GVSYY831 pKa = 10.26EE832 pKa = 4.36SNSMLTALASASDD845 pKa = 4.17PDD847 pKa = 4.19GEE849 pKa = 4.16IANVSFYY856 pKa = 10.52MDD858 pKa = 3.44GNEE861 pKa = 4.21IGSATSSPYY870 pKa = 10.23EE871 pKa = 3.91YY872 pKa = 10.44EE873 pKa = 3.89IDD875 pKa = 3.54LSIYY879 pKa = 10.27EE880 pKa = 4.15LGSYY884 pKa = 9.91EE885 pKa = 4.06FFAIAEE891 pKa = 4.52DD892 pKa = 4.29DD893 pKa = 4.32DD894 pKa = 4.58GALASSDD901 pKa = 3.37TVTFEE906 pKa = 4.36IIEE909 pKa = 4.41GNGGGSEE916 pKa = 4.03LPARR920 pKa = 11.84VMEE923 pKa = 5.34DD924 pKa = 2.7IDD926 pKa = 4.26RR927 pKa = 11.84GLIAIDD933 pKa = 4.03RR934 pKa = 11.84GDD936 pKa = 4.98DD937 pKa = 3.4IYY939 pKa = 11.52LSWRR943 pKa = 11.84LLGKK947 pKa = 8.89EE948 pKa = 3.98APDD951 pKa = 3.59LGFNIYY957 pKa = 10.22RR958 pKa = 11.84DD959 pKa = 3.69GTLVNSEE966 pKa = 5.0PITDD970 pKa = 3.44RR971 pKa = 11.84TNYY974 pKa = 9.86VDD976 pKa = 4.6ADD978 pKa = 3.76GSADD982 pKa = 3.38ANYY985 pKa = 10.9SVIPVEE991 pKa = 5.27DD992 pKa = 3.63GTEE995 pKa = 3.97LSEE998 pKa = 4.6SEE1000 pKa = 3.99ATASVWANSFIDD1012 pKa = 3.42IPLNIPADD1020 pKa = 3.93GTHH1023 pKa = 6.4GGSYY1027 pKa = 10.31SPNDD1031 pKa = 3.59MSIGDD1036 pKa = 4.07LDD1038 pKa = 3.95GDD1040 pKa = 3.88NEE1042 pKa = 4.34YY1043 pKa = 11.23EE1044 pKa = 5.87LVLKK1048 pKa = 9.36WYY1050 pKa = 9.32PSNAKK1055 pKa = 10.5DD1056 pKa = 3.52NAQDD1060 pKa = 3.49GLTDD1064 pKa = 3.5QTILEE1069 pKa = 4.19GLEE1072 pKa = 4.04LDD1074 pKa = 4.05GTSMWRR1080 pKa = 11.84IEE1082 pKa = 4.2LGDD1085 pKa = 3.9NIRR1088 pKa = 11.84SGAHH1092 pKa = 3.63YY1093 pKa = 8.61TQFIVYY1099 pKa = 10.26DD1100 pKa = 4.12FDD1102 pKa = 5.33SDD1104 pKa = 3.78GKK1106 pKa = 11.17AEE1108 pKa = 4.27VALKK1112 pKa = 8.17TAPGTRR1118 pKa = 11.84DD1119 pKa = 3.04GEE1121 pKa = 4.5GNYY1124 pKa = 10.53LSTGPAADD1132 pKa = 4.94DD1133 pKa = 4.66DD1134 pKa = 4.68DD1135 pKa = 4.39AASYY1139 pKa = 7.72VTNTGRR1145 pKa = 11.84IIYY1148 pKa = 7.25PSPEE1152 pKa = 3.81YY1153 pKa = 10.23LTIFEE1158 pKa = 4.49GATGKK1163 pKa = 9.89EE1164 pKa = 4.29LQTEE1168 pKa = 5.14LYY1170 pKa = 10.17SPQYY1174 pKa = 10.6DD1175 pKa = 3.6FKK1177 pKa = 11.72NNPEE1181 pKa = 4.12NFWGDD1186 pKa = 3.38GYY1188 pKa = 11.15GNRR1191 pKa = 11.84SEE1193 pKa = 5.26RR1194 pKa = 11.84YY1195 pKa = 8.11LAAVGYY1201 pKa = 9.94FNGEE1205 pKa = 4.08SPSLVMSRR1213 pKa = 11.84GYY1215 pKa = 10.56YY1216 pKa = 9.13EE1217 pKa = 5.07GYY1219 pKa = 10.37VVAAWDD1225 pKa = 3.48WDD1227 pKa = 3.79GEE1229 pKa = 4.28NLTNRR1234 pKa = 11.84WNFEE1238 pKa = 4.03AGPWSSDD1245 pKa = 3.12PYY1247 pKa = 10.78GGQGNHH1253 pKa = 6.65NIAVGDD1259 pKa = 3.63VDD1261 pKa = 5.13EE1262 pKa = 5.46DD1263 pKa = 4.01GKK1265 pKa = 11.02DD1266 pKa = 3.33EE1267 pKa = 4.68IMFGSAAIDD1276 pKa = 3.71DD1277 pKa = 5.25DD1278 pKa = 4.23GTGLYY1283 pKa = 6.72TTRR1286 pKa = 11.84NGHH1289 pKa = 6.07GDD1291 pKa = 3.48AGHH1294 pKa = 7.36LGDD1297 pKa = 5.85LDD1299 pKa = 4.04PDD1301 pKa = 3.57IPGLEE1306 pKa = 3.68FFMPHH1311 pKa = 5.59EE1312 pKa = 4.12WSGPGLSFRR1321 pKa = 11.84NAGTGSMIWEE1331 pKa = 4.51VPSDD1335 pKa = 3.81DD1336 pKa = 4.02DD1337 pKa = 3.8VGRR1340 pKa = 11.84GVAEE1344 pKa = 5.27DD1345 pKa = 3.49ISAEE1349 pKa = 3.96YY1350 pKa = 10.22RR1351 pKa = 11.84GAEE1354 pKa = 3.71AWGLGAYY1361 pKa = 7.36NAKK1364 pKa = 10.08GDD1366 pKa = 4.92PIDD1369 pKa = 5.11LSPPSANFVIYY1380 pKa = 9.84WDD1382 pKa = 3.72GDD1384 pKa = 3.69VTRR1387 pKa = 11.84EE1388 pKa = 4.0LLDD1391 pKa = 3.48RR1392 pKa = 11.84NYY1394 pKa = 10.11IDD1396 pKa = 5.28DD1397 pKa = 4.22MEE1399 pKa = 4.64NGRR1402 pKa = 11.84IFTADD1407 pKa = 2.95GFSSNNGTKK1416 pKa = 8.93ATPNLSVDD1424 pKa = 4.19LFGDD1428 pKa = 2.98WRR1430 pKa = 11.84EE1431 pKa = 4.01EE1432 pKa = 4.07VIFRR1436 pKa = 11.84SDD1438 pKa = 3.59DD1439 pKa = 3.53NTSLRR1444 pKa = 11.84IFTTSDD1450 pKa = 2.69TSDD1453 pKa = 2.91VRR1455 pKa = 11.84RR1456 pKa = 11.84YY1457 pKa = 8.44TLMHH1461 pKa = 7.02DD1462 pKa = 3.27PLYY1465 pKa = 10.26RR1466 pKa = 11.84ISIAWQNVAYY1476 pKa = 9.28NQPPHH1481 pKa = 6.88VGYY1484 pKa = 10.93YY1485 pKa = 9.17MPDD1488 pKa = 3.46GSPQPNIIYY1497 pKa = 9.03PDD1499 pKa = 3.65EE1500 pKa = 4.26YY1501 pKa = 10.96VSVSINPEE1509 pKa = 3.84GKK1511 pKa = 9.86DD1512 pKa = 3.28GSVIKK1517 pKa = 10.22DD1518 pKa = 3.45YY1519 pKa = 11.3KK1520 pKa = 9.21LQNYY1524 pKa = 7.31PNPFNPTTNISFSLPVSGDD1543 pKa = 3.1VKK1545 pKa = 11.35LEE1547 pKa = 3.84VFDD1550 pKa = 3.89INGRR1554 pKa = 11.84LISTIVNEE1562 pKa = 4.18RR1563 pKa = 11.84MTSGTYY1569 pKa = 10.34SRR1571 pKa = 11.84TFDD1574 pKa = 3.57ASNLASGIYY1583 pKa = 10.11FYY1585 pKa = 11.31RR1586 pKa = 11.84LRR1588 pKa = 11.84TSDD1591 pKa = 3.41YY1592 pKa = 8.54TQVKK1596 pKa = 10.5KK1597 pKa = 10.82MMLIKK1602 pKa = 10.65
MM1 pKa = 7.81GIFPYY6 pKa = 10.55AQAQEE11 pKa = 4.36VVTTFDD17 pKa = 4.39LNGADD22 pKa = 3.11GTLYY26 pKa = 10.77NDD28 pKa = 3.91EE29 pKa = 4.29NRR31 pKa = 11.84GADD34 pKa = 3.62FPGGDD39 pKa = 3.42RR40 pKa = 11.84SSLEE44 pKa = 3.14IRR46 pKa = 11.84YY47 pKa = 8.55WDD49 pKa = 3.45EE50 pKa = 3.6VRR52 pKa = 11.84FRR54 pKa = 11.84ASMVRR59 pKa = 11.84FDD61 pKa = 3.93VNAVNSSPDD70 pKa = 3.41GAVLGLYY77 pKa = 6.87PTSAGSMGADD87 pKa = 2.53SLTFTIYY94 pKa = 11.2GLTNEE99 pKa = 5.67SEE101 pKa = 4.7DD102 pKa = 3.49NWDD105 pKa = 3.74EE106 pKa = 4.37GSFSYY111 pKa = 11.3NEE113 pKa = 3.94APGFEE118 pKa = 4.04AAANGTYY125 pKa = 10.29AINEE129 pKa = 4.35DD130 pKa = 4.4LDD132 pKa = 3.9SLTTITVYY140 pKa = 11.35NNLTDD145 pKa = 3.44TYY147 pKa = 10.66LYY149 pKa = 10.47SEE151 pKa = 5.19PSAEE155 pKa = 4.21LDD157 pKa = 3.42AFINSDD163 pKa = 3.67TNGLVTFAIVKK174 pKa = 8.7TYY176 pKa = 8.63STEE179 pKa = 3.77SGSYY183 pKa = 9.91VISPKK188 pKa = 10.36EE189 pKa = 3.86AGEE192 pKa = 4.28SVAPKK197 pKa = 10.26LVYY200 pKa = 10.12QSEE203 pKa = 4.32NGEE206 pKa = 3.88PVLPNTEE213 pKa = 3.85PSITLDD219 pKa = 3.53SPTDD223 pKa = 3.59GASYY227 pKa = 10.59EE228 pKa = 4.52SNNTLTASASASDD241 pKa = 3.82SDD243 pKa = 4.64GEE245 pKa = 4.11IASVSFYY252 pKa = 11.17LDD254 pKa = 3.19DD255 pKa = 4.0TEE257 pKa = 5.04IGSVNASPFEE267 pKa = 4.0YY268 pKa = 10.47EE269 pKa = 4.31IDD271 pKa = 3.41LSMYY275 pKa = 11.01ALGSHH280 pKa = 6.47EE281 pKa = 4.84FYY283 pKa = 10.85AIAEE287 pKa = 4.53DD288 pKa = 3.58NDD290 pKa = 4.01GAVASSDD297 pKa = 2.95TVMFEE302 pKa = 3.34ITEE305 pKa = 4.74AIGGSDD311 pKa = 3.57AEE313 pKa = 4.32MVTTFEE319 pKa = 5.57LNGADD324 pKa = 3.21AAIYY328 pKa = 10.86NDD330 pKa = 4.0DD331 pKa = 4.2NRR333 pKa = 11.84GADD336 pKa = 3.46YY337 pKa = 10.84TGGNRR342 pKa = 11.84SDD344 pKa = 3.97LEE346 pKa = 3.55IRR348 pKa = 11.84YY349 pKa = 8.63WEE351 pKa = 4.1EE352 pKa = 3.3VRR354 pKa = 11.84FRR356 pKa = 11.84ASILRR361 pKa = 11.84FDD363 pKa = 4.01VSNVSSTPNGAVIGFNPTYY382 pKa = 9.47AGKK385 pKa = 9.24MGAEE389 pKa = 4.06SLTYY393 pKa = 9.73TIYY396 pKa = 11.33GMTDD400 pKa = 3.17EE401 pKa = 5.82SQDD404 pKa = 3.28NWDD407 pKa = 4.08EE408 pKa = 4.17GSVNYY413 pKa = 10.46SSAPGFEE420 pKa = 4.36SAANGTYY427 pKa = 11.19ALTEE431 pKa = 4.14NMDD434 pKa = 3.96SLTTITVYY442 pKa = 11.15ADD444 pKa = 3.02STGAFLYY451 pKa = 10.48SDD453 pKa = 4.62PSTEE457 pKa = 4.2LDD459 pKa = 3.22AFINNDD465 pKa = 3.25SNGLVTFAIIKK476 pKa = 8.02TISTNSDD483 pKa = 3.64YY484 pKa = 11.66YY485 pKa = 11.89AMNSKK490 pKa = 10.02EE491 pKa = 4.18AGVSVAPRR499 pKa = 11.84LVYY502 pKa = 10.33QSEE505 pKa = 4.15NGEE508 pKa = 3.83PVLPNFEE515 pKa = 4.23PSITLNSPEE524 pKa = 4.31EE525 pKa = 4.11GMVYY529 pKa = 10.43QSDD532 pKa = 3.83STLTARR538 pKa = 11.84ATAIDD543 pKa = 3.86SDD545 pKa = 4.59GEE547 pKa = 4.04IASVSFYY554 pKa = 10.55MDD556 pKa = 3.52GTAVANVTSSPYY568 pKa = 10.15EE569 pKa = 4.01SEE571 pKa = 4.32IDD573 pKa = 3.36LSMYY577 pKa = 11.03ASGSYY582 pKa = 9.67EE583 pKa = 3.93FYY585 pKa = 10.83AIAEE589 pKa = 4.59DD590 pKa = 4.87DD591 pKa = 4.46DD592 pKa = 4.59GATASSDD599 pKa = 3.25TVTIDD604 pKa = 2.94IVGGSGAGSSEE615 pKa = 3.94MVTTFDD621 pKa = 4.66SNGADD626 pKa = 2.97VTLFNDD632 pKa = 3.55EE633 pKa = 3.94NRR635 pKa = 11.84AGSFTGGDD643 pKa = 3.23RR644 pKa = 11.84TDD646 pKa = 3.56LEE648 pKa = 4.06VRR650 pKa = 11.84YY651 pKa = 9.19WEE653 pKa = 4.03AVRR656 pKa = 11.84FRR658 pKa = 11.84ASMLRR663 pKa = 11.84FDD665 pKa = 4.29VSSVSAAPDD674 pKa = 3.46GAVIGINPTYY684 pKa = 10.4AEE686 pKa = 4.07NMSDD690 pKa = 3.29EE691 pKa = 4.5TLSFTIFGLTNEE703 pKa = 5.5AEE705 pKa = 4.42DD706 pKa = 3.78EE707 pKa = 3.99WDD709 pKa = 3.72EE710 pKa = 4.8GSISYY715 pKa = 10.81NSAPGFEE722 pKa = 4.45SAINGSYY729 pKa = 10.59AINDD733 pKa = 4.15DD734 pKa = 4.36LDD736 pKa = 3.82SLTTITVYY744 pKa = 11.14ADD746 pKa = 3.13STGTFLYY753 pKa = 10.57SEE755 pKa = 4.96PSDD758 pKa = 3.75KK759 pKa = 11.07LDD761 pKa = 3.72AFIRR765 pKa = 11.84NDD767 pKa = 3.23NNGLVTFAIIRR778 pKa = 11.84TYY780 pKa = 11.43SNNSDD785 pKa = 3.18IYY787 pKa = 11.42AMSSKK792 pKa = 10.42EE793 pKa = 3.87AGEE796 pKa = 4.01LVAPRR801 pKa = 11.84LVYY804 pKa = 10.38KK805 pKa = 10.5SEE807 pKa = 4.04NGVPVIPNLEE817 pKa = 4.1PNITLSRR824 pKa = 11.84PADD827 pKa = 3.57GVSYY831 pKa = 10.26EE832 pKa = 4.36SNSMLTALASASDD845 pKa = 4.17PDD847 pKa = 4.19GEE849 pKa = 4.16IANVSFYY856 pKa = 10.52MDD858 pKa = 3.44GNEE861 pKa = 4.21IGSATSSPYY870 pKa = 10.23EE871 pKa = 3.91YY872 pKa = 10.44EE873 pKa = 3.89IDD875 pKa = 3.54LSIYY879 pKa = 10.27EE880 pKa = 4.15LGSYY884 pKa = 9.91EE885 pKa = 4.06FFAIAEE891 pKa = 4.52DD892 pKa = 4.29DD893 pKa = 4.32DD894 pKa = 4.58GALASSDD901 pKa = 3.37TVTFEE906 pKa = 4.36IIEE909 pKa = 4.41GNGGGSEE916 pKa = 4.03LPARR920 pKa = 11.84VMEE923 pKa = 5.34DD924 pKa = 2.7IDD926 pKa = 4.26RR927 pKa = 11.84GLIAIDD933 pKa = 4.03RR934 pKa = 11.84GDD936 pKa = 4.98DD937 pKa = 3.4IYY939 pKa = 11.52LSWRR943 pKa = 11.84LLGKK947 pKa = 8.89EE948 pKa = 3.98APDD951 pKa = 3.59LGFNIYY957 pKa = 10.22RR958 pKa = 11.84DD959 pKa = 3.69GTLVNSEE966 pKa = 5.0PITDD970 pKa = 3.44RR971 pKa = 11.84TNYY974 pKa = 9.86VDD976 pKa = 4.6ADD978 pKa = 3.76GSADD982 pKa = 3.38ANYY985 pKa = 10.9SVIPVEE991 pKa = 5.27DD992 pKa = 3.63GTEE995 pKa = 3.97LSEE998 pKa = 4.6SEE1000 pKa = 3.99ATASVWANSFIDD1012 pKa = 3.42IPLNIPADD1020 pKa = 3.93GTHH1023 pKa = 6.4GGSYY1027 pKa = 10.31SPNDD1031 pKa = 3.59MSIGDD1036 pKa = 4.07LDD1038 pKa = 3.95GDD1040 pKa = 3.88NEE1042 pKa = 4.34YY1043 pKa = 11.23EE1044 pKa = 5.87LVLKK1048 pKa = 9.36WYY1050 pKa = 9.32PSNAKK1055 pKa = 10.5DD1056 pKa = 3.52NAQDD1060 pKa = 3.49GLTDD1064 pKa = 3.5QTILEE1069 pKa = 4.19GLEE1072 pKa = 4.04LDD1074 pKa = 4.05GTSMWRR1080 pKa = 11.84IEE1082 pKa = 4.2LGDD1085 pKa = 3.9NIRR1088 pKa = 11.84SGAHH1092 pKa = 3.63YY1093 pKa = 8.61TQFIVYY1099 pKa = 10.26DD1100 pKa = 4.12FDD1102 pKa = 5.33SDD1104 pKa = 3.78GKK1106 pKa = 11.17AEE1108 pKa = 4.27VALKK1112 pKa = 8.17TAPGTRR1118 pKa = 11.84DD1119 pKa = 3.04GEE1121 pKa = 4.5GNYY1124 pKa = 10.53LSTGPAADD1132 pKa = 4.94DD1133 pKa = 4.66DD1134 pKa = 4.68DD1135 pKa = 4.39AASYY1139 pKa = 7.72VTNTGRR1145 pKa = 11.84IIYY1148 pKa = 7.25PSPEE1152 pKa = 3.81YY1153 pKa = 10.23LTIFEE1158 pKa = 4.49GATGKK1163 pKa = 9.89EE1164 pKa = 4.29LQTEE1168 pKa = 5.14LYY1170 pKa = 10.17SPQYY1174 pKa = 10.6DD1175 pKa = 3.6FKK1177 pKa = 11.72NNPEE1181 pKa = 4.12NFWGDD1186 pKa = 3.38GYY1188 pKa = 11.15GNRR1191 pKa = 11.84SEE1193 pKa = 5.26RR1194 pKa = 11.84YY1195 pKa = 8.11LAAVGYY1201 pKa = 9.94FNGEE1205 pKa = 4.08SPSLVMSRR1213 pKa = 11.84GYY1215 pKa = 10.56YY1216 pKa = 9.13EE1217 pKa = 5.07GYY1219 pKa = 10.37VVAAWDD1225 pKa = 3.48WDD1227 pKa = 3.79GEE1229 pKa = 4.28NLTNRR1234 pKa = 11.84WNFEE1238 pKa = 4.03AGPWSSDD1245 pKa = 3.12PYY1247 pKa = 10.78GGQGNHH1253 pKa = 6.65NIAVGDD1259 pKa = 3.63VDD1261 pKa = 5.13EE1262 pKa = 5.46DD1263 pKa = 4.01GKK1265 pKa = 11.02DD1266 pKa = 3.33EE1267 pKa = 4.68IMFGSAAIDD1276 pKa = 3.71DD1277 pKa = 5.25DD1278 pKa = 4.23GTGLYY1283 pKa = 6.72TTRR1286 pKa = 11.84NGHH1289 pKa = 6.07GDD1291 pKa = 3.48AGHH1294 pKa = 7.36LGDD1297 pKa = 5.85LDD1299 pKa = 4.04PDD1301 pKa = 3.57IPGLEE1306 pKa = 3.68FFMPHH1311 pKa = 5.59EE1312 pKa = 4.12WSGPGLSFRR1321 pKa = 11.84NAGTGSMIWEE1331 pKa = 4.51VPSDD1335 pKa = 3.81DD1336 pKa = 4.02DD1337 pKa = 3.8VGRR1340 pKa = 11.84GVAEE1344 pKa = 5.27DD1345 pKa = 3.49ISAEE1349 pKa = 3.96YY1350 pKa = 10.22RR1351 pKa = 11.84GAEE1354 pKa = 3.71AWGLGAYY1361 pKa = 7.36NAKK1364 pKa = 10.08GDD1366 pKa = 4.92PIDD1369 pKa = 5.11LSPPSANFVIYY1380 pKa = 9.84WDD1382 pKa = 3.72GDD1384 pKa = 3.69VTRR1387 pKa = 11.84EE1388 pKa = 4.0LLDD1391 pKa = 3.48RR1392 pKa = 11.84NYY1394 pKa = 10.11IDD1396 pKa = 5.28DD1397 pKa = 4.22MEE1399 pKa = 4.64NGRR1402 pKa = 11.84IFTADD1407 pKa = 2.95GFSSNNGTKK1416 pKa = 8.93ATPNLSVDD1424 pKa = 4.19LFGDD1428 pKa = 2.98WRR1430 pKa = 11.84EE1431 pKa = 4.01EE1432 pKa = 4.07VIFRR1436 pKa = 11.84SDD1438 pKa = 3.59DD1439 pKa = 3.53NTSLRR1444 pKa = 11.84IFTTSDD1450 pKa = 2.69TSDD1453 pKa = 2.91VRR1455 pKa = 11.84RR1456 pKa = 11.84YY1457 pKa = 8.44TLMHH1461 pKa = 7.02DD1462 pKa = 3.27PLYY1465 pKa = 10.26RR1466 pKa = 11.84ISIAWQNVAYY1476 pKa = 9.28NQPPHH1481 pKa = 6.88VGYY1484 pKa = 10.93YY1485 pKa = 9.17MPDD1488 pKa = 3.46GSPQPNIIYY1497 pKa = 9.03PDD1499 pKa = 3.65EE1500 pKa = 4.26YY1501 pKa = 10.96VSVSINPEE1509 pKa = 3.84GKK1511 pKa = 9.86DD1512 pKa = 3.28GSVIKK1517 pKa = 10.22DD1518 pKa = 3.45YY1519 pKa = 11.3KK1520 pKa = 9.21LQNYY1524 pKa = 7.31PNPFNPTTNISFSLPVSGDD1543 pKa = 3.1VKK1545 pKa = 11.35LEE1547 pKa = 3.84VFDD1550 pKa = 3.89INGRR1554 pKa = 11.84LISTIVNEE1562 pKa = 4.18RR1563 pKa = 11.84MTSGTYY1569 pKa = 10.34SRR1571 pKa = 11.84TFDD1574 pKa = 3.57ASNLASGIYY1583 pKa = 10.11FYY1585 pKa = 11.31RR1586 pKa = 11.84LRR1588 pKa = 11.84TSDD1591 pKa = 3.41YY1592 pKa = 8.54TQVKK1596 pKa = 10.5KK1597 pKa = 10.82MMLIKK1602 pKa = 10.65
Molecular weight: 173.84 kDa
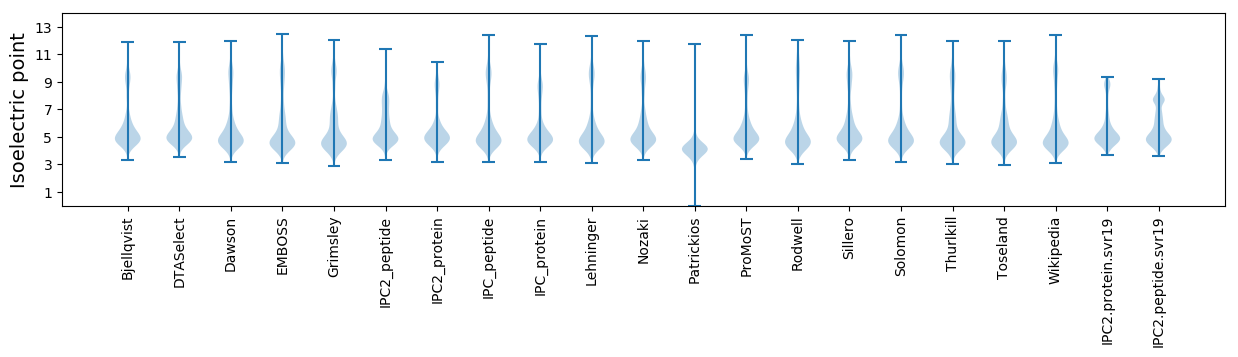
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A521CVQ6|A0A521CVQ6_9BACT Uncharacterized protein OS=Gracilimonas mengyeensis OX=1302730 GN=SAMN06265219_106133 PE=4 SV=1
MM1 pKa = 7.89ADD3 pKa = 3.27YY4 pKa = 10.97SHH6 pKa = 7.31RR7 pKa = 11.84FPIRR11 pKa = 11.84KK12 pKa = 7.63MSQVFGVSPSGYY24 pKa = 8.93YY25 pKa = 9.45RR26 pKa = 11.84WLGRR30 pKa = 11.84PPSKK34 pKa = 10.17RR35 pKa = 11.84ARR37 pKa = 11.84QNRR40 pKa = 11.84RR41 pKa = 11.84LKK43 pKa = 10.64EE44 pKa = 4.14SIRR47 pKa = 11.84LVWVLSKK54 pKa = 10.57RR55 pKa = 11.84IYY57 pKa = 9.75GAPRR61 pKa = 11.84IYY63 pKa = 10.42QEE65 pKa = 3.85LLRR68 pKa = 11.84RR69 pKa = 11.84GWQVSRR75 pKa = 11.84PRR77 pKa = 11.84VARR80 pKa = 11.84LMRR83 pKa = 11.84KK84 pKa = 8.53MGISSQLRR92 pKa = 11.84KK93 pKa = 10.0KK94 pKa = 8.34WVKK97 pKa = 7.05TTNSKK102 pKa = 10.26HH103 pKa = 4.48SHH105 pKa = 5.62PVAGNLLDD113 pKa = 4.61RR114 pKa = 11.84RR115 pKa = 11.84FTPTQLNQVWVSDD128 pKa = 3.38ITYY131 pKa = 10.32LPSQQGWLYY140 pKa = 9.11LTTVMDD146 pKa = 4.43LADD149 pKa = 4.09RR150 pKa = 11.84QILGWSLSTSMSAEE164 pKa = 3.93QTTTAAFCEE173 pKa = 4.32AHH175 pKa = 6.05TKK177 pKa = 10.22RR178 pKa = 11.84PARR181 pKa = 11.84KK182 pKa = 9.9GMIFHH187 pKa = 7.45SDD189 pKa = 2.44RR190 pKa = 11.84GMQYY194 pKa = 10.84ACSAFTNLLSKK205 pKa = 10.47HH206 pKa = 5.59QLIQSMSRR214 pKa = 11.84KK215 pKa = 9.17GNCWDD220 pKa = 3.56NAPAEE225 pKa = 4.59SFFKK229 pKa = 10.17TLKK232 pKa = 10.86AEE234 pKa = 4.28LVSQTGIFRR243 pKa = 11.84DD244 pKa = 3.93YY245 pKa = 10.17QQARR249 pKa = 11.84AAIFEE254 pKa = 4.64YY255 pKa = 10.6IEE257 pKa = 3.51GWYY260 pKa = 9.3NRR262 pKa = 11.84KK263 pKa = 9.23RR264 pKa = 11.84LHH266 pKa = 6.77SSLGYY271 pKa = 8.09KK272 pKa = 9.29TPAEE276 pKa = 4.33AEE278 pKa = 4.02QEE280 pKa = 4.22LKK282 pKa = 10.78QKK284 pKa = 10.3LQQAAA289 pKa = 3.29
MM1 pKa = 7.89ADD3 pKa = 3.27YY4 pKa = 10.97SHH6 pKa = 7.31RR7 pKa = 11.84FPIRR11 pKa = 11.84KK12 pKa = 7.63MSQVFGVSPSGYY24 pKa = 8.93YY25 pKa = 9.45RR26 pKa = 11.84WLGRR30 pKa = 11.84PPSKK34 pKa = 10.17RR35 pKa = 11.84ARR37 pKa = 11.84QNRR40 pKa = 11.84RR41 pKa = 11.84LKK43 pKa = 10.64EE44 pKa = 4.14SIRR47 pKa = 11.84LVWVLSKK54 pKa = 10.57RR55 pKa = 11.84IYY57 pKa = 9.75GAPRR61 pKa = 11.84IYY63 pKa = 10.42QEE65 pKa = 3.85LLRR68 pKa = 11.84RR69 pKa = 11.84GWQVSRR75 pKa = 11.84PRR77 pKa = 11.84VARR80 pKa = 11.84LMRR83 pKa = 11.84KK84 pKa = 8.53MGISSQLRR92 pKa = 11.84KK93 pKa = 10.0KK94 pKa = 8.34WVKK97 pKa = 7.05TTNSKK102 pKa = 10.26HH103 pKa = 4.48SHH105 pKa = 5.62PVAGNLLDD113 pKa = 4.61RR114 pKa = 11.84RR115 pKa = 11.84FTPTQLNQVWVSDD128 pKa = 3.38ITYY131 pKa = 10.32LPSQQGWLYY140 pKa = 9.11LTTVMDD146 pKa = 4.43LADD149 pKa = 4.09RR150 pKa = 11.84QILGWSLSTSMSAEE164 pKa = 3.93QTTTAAFCEE173 pKa = 4.32AHH175 pKa = 6.05TKK177 pKa = 10.22RR178 pKa = 11.84PARR181 pKa = 11.84KK182 pKa = 9.9GMIFHH187 pKa = 7.45SDD189 pKa = 2.44RR190 pKa = 11.84GMQYY194 pKa = 10.84ACSAFTNLLSKK205 pKa = 10.47HH206 pKa = 5.59QLIQSMSRR214 pKa = 11.84KK215 pKa = 9.17GNCWDD220 pKa = 3.56NAPAEE225 pKa = 4.59SFFKK229 pKa = 10.17TLKK232 pKa = 10.86AEE234 pKa = 4.28LVSQTGIFRR243 pKa = 11.84DD244 pKa = 3.93YY245 pKa = 10.17QQARR249 pKa = 11.84AAIFEE254 pKa = 4.64YY255 pKa = 10.6IEE257 pKa = 3.51GWYY260 pKa = 9.3NRR262 pKa = 11.84KK263 pKa = 9.23RR264 pKa = 11.84LHH266 pKa = 6.77SSLGYY271 pKa = 8.09KK272 pKa = 9.29TPAEE276 pKa = 4.33AEE278 pKa = 4.02QEE280 pKa = 4.22LKK282 pKa = 10.78QKK284 pKa = 10.3LQQAAA289 pKa = 3.29
Molecular weight: 33.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1339685 |
24 |
4477 |
351.6 |
39.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.06 ± 0.035 | 0.553 ± 0.01 |
6.057 ± 0.031 | 7.777 ± 0.047 |
4.72 ± 0.028 | 7.165 ± 0.036 |
1.933 ± 0.02 | 6.672 ± 0.033 |
5.607 ± 0.052 | 9.183 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.419 ± 0.021 | 4.858 ± 0.033 |
3.95 ± 0.022 | 3.829 ± 0.025 |
4.371 ± 0.03 | 6.648 ± 0.038 |
5.586 ± 0.043 | 6.499 ± 0.033 |
1.298 ± 0.017 | 3.816 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
