
Corallincola sp. C4
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Psychromonadaceae; Corallincola
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
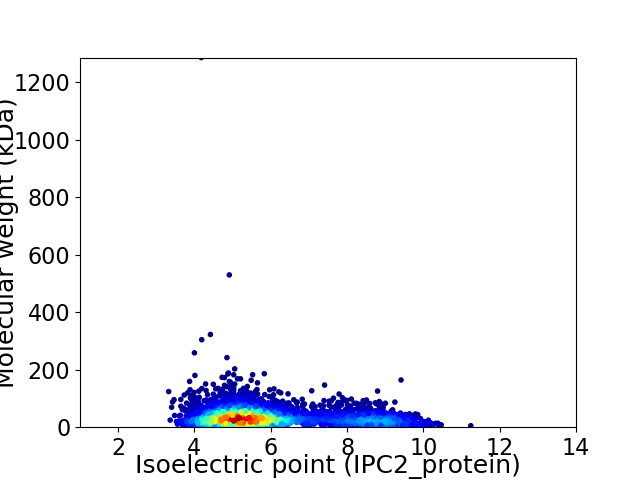
Virtual 2D-PAGE plot for 3747 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A368NJE5|A0A368NJE5_9GAMM Hotdog fold thioesterase OS=Corallincola sp. C4 OX=2282215 GN=DU002_10280 PE=4 SV=1
MM1 pKa = 7.82RR2 pKa = 11.84KK3 pKa = 10.1LIITLAVASALAVTGCDD20 pKa = 5.25DD21 pKa = 4.69EE22 pKa = 5.73SLDD25 pKa = 5.24DD26 pKa = 4.69ARR28 pKa = 11.84NDD30 pKa = 3.58AAGTPVINASKK41 pKa = 10.35ISYY44 pKa = 10.23DD45 pKa = 3.48PANGVLPVPNDD56 pKa = 3.67LLYY59 pKa = 10.69TDD61 pKa = 4.24TTDD64 pKa = 3.02GTLNIPVTDD73 pKa = 4.67PSDD76 pKa = 3.42LSNPQVAINALDD88 pKa = 3.51GWSTNQPFVINVITDD103 pKa = 3.31GDD105 pKa = 4.06VTLDD109 pKa = 3.82PASAAAPGVVRR120 pKa = 11.84IYY122 pKa = 9.22EE123 pKa = 4.38TVFGGPLAPPEE134 pKa = 4.26CAEE137 pKa = 4.27VPTGAACMVIAEE149 pKa = 4.78LAFGTDD155 pKa = 5.05FITSSSGNQLLIMPLAPLKK174 pKa = 10.99AKK176 pKa = 9.37TGYY179 pKa = 10.93LITTTSLLMDD189 pKa = 3.82SEE191 pKa = 4.46SRR193 pKa = 11.84PVQASSTYY201 pKa = 9.99TLVKK205 pKa = 10.24QDD207 pKa = 3.99INTLPLGTPSQLLLQGAVNSYY228 pKa = 10.11EE229 pKa = 3.85NAIAAFGVDD238 pKa = 3.48TSVITYY244 pKa = 9.49SGAMTTQSIEE254 pKa = 4.18DD255 pKa = 3.47VSALNRR261 pKa = 11.84LTMAADD267 pKa = 3.77PATTPVLMPPAMTGLTVADD286 pKa = 4.08ALYY289 pKa = 10.65EE290 pKa = 4.07LGIRR294 pKa = 11.84DD295 pKa = 4.22TNTLAVASTALLANSTLTNVPYY317 pKa = 10.69YY318 pKa = 11.04LEE320 pKa = 4.51FPNFGNCDD328 pKa = 3.55LTEE331 pKa = 4.29LATTSQCDD339 pKa = 3.64AINSHH344 pKa = 5.96WLAAGDD350 pKa = 4.07SPVTILGAVQSGALPPEE367 pKa = 4.48AVLASCPDD375 pKa = 3.67ADD377 pKa = 4.62LSNAASLVGCEE388 pKa = 4.3IKK390 pKa = 10.63DD391 pKa = 3.7AEE393 pKa = 4.49GNPIGLDD400 pKa = 3.3PEE402 pKa = 4.78RR403 pKa = 11.84NLTKK407 pKa = 10.5YY408 pKa = 10.64NPLAAPTEE416 pKa = 4.21MQTLDD421 pKa = 3.68VLVTLPDD428 pKa = 3.3TSDD431 pKa = 3.06AANGVRR437 pKa = 11.84AALGLPAITAAPAAGWPVVVFSHH460 pKa = 7.18GIGGAKK466 pKa = 8.83EE467 pKa = 3.88QNLALAGSFALQGLAMVSIDD487 pKa = 3.88APLHH491 pKa = 5.84GSRR494 pKa = 11.84GWDD497 pKa = 2.78ADD499 pKa = 3.62MNGVFEE505 pKa = 4.87ISASSGMIAFDD516 pKa = 3.42PVKK519 pKa = 10.39YY520 pKa = 10.88ANANVLLYY528 pKa = 11.0VKK530 pKa = 10.37LDD532 pKa = 3.61NLLTTRR538 pKa = 11.84DD539 pKa = 3.53NFRR542 pKa = 11.84QNINDD547 pKa = 3.73HH548 pKa = 6.17LALRR552 pKa = 11.84MGFNSLAQVAPGVFDD567 pKa = 3.58PTRR570 pKa = 11.84VSISGVSLGGINTVSTAAVGGLPLIDD596 pKa = 3.86PTTGSEE602 pKa = 4.19LPSYY606 pKa = 10.24YY607 pKa = 10.72AFSTVMPNVPAQGLAGVFAYY627 pKa = 10.47SEE629 pKa = 4.43TFGPVAKK636 pKa = 10.27AAFVEE641 pKa = 4.35SDD643 pKa = 3.31SFKK646 pKa = 11.0QVLSDD651 pKa = 3.46ATGLSLEE658 pKa = 4.22EE659 pKa = 4.01LAALEE664 pKa = 4.82ASDD667 pKa = 3.97PDD669 pKa = 3.86AYY671 pKa = 10.48QALVDD676 pKa = 3.67QVYY679 pKa = 8.14PTFIAQFVFAAQTVVDD695 pKa = 3.87SSDD698 pKa = 3.44PVGYY702 pKa = 10.77APILAGQGVPVYY714 pKa = 9.94LAEE717 pKa = 4.31VVGDD721 pKa = 3.63TVLPNDD727 pKa = 3.52NSAFGLPLSGTEE739 pKa = 3.61PLIRR743 pKa = 11.84VLGLPGVDD751 pKa = 3.14ATVTGAPATGAVRR764 pKa = 11.84FIDD767 pKa = 3.78SAHH770 pKa = 6.54GSVADD775 pKa = 3.72PTVNPAATVEE785 pKa = 4.31MQTQLAVYY793 pKa = 8.5AASGGAMILVSDD805 pKa = 4.59PSVVQPVMM813 pKa = 3.63
MM1 pKa = 7.82RR2 pKa = 11.84KK3 pKa = 10.1LIITLAVASALAVTGCDD20 pKa = 5.25DD21 pKa = 4.69EE22 pKa = 5.73SLDD25 pKa = 5.24DD26 pKa = 4.69ARR28 pKa = 11.84NDD30 pKa = 3.58AAGTPVINASKK41 pKa = 10.35ISYY44 pKa = 10.23DD45 pKa = 3.48PANGVLPVPNDD56 pKa = 3.67LLYY59 pKa = 10.69TDD61 pKa = 4.24TTDD64 pKa = 3.02GTLNIPVTDD73 pKa = 4.67PSDD76 pKa = 3.42LSNPQVAINALDD88 pKa = 3.51GWSTNQPFVINVITDD103 pKa = 3.31GDD105 pKa = 4.06VTLDD109 pKa = 3.82PASAAAPGVVRR120 pKa = 11.84IYY122 pKa = 9.22EE123 pKa = 4.38TVFGGPLAPPEE134 pKa = 4.26CAEE137 pKa = 4.27VPTGAACMVIAEE149 pKa = 4.78LAFGTDD155 pKa = 5.05FITSSSGNQLLIMPLAPLKK174 pKa = 10.99AKK176 pKa = 9.37TGYY179 pKa = 10.93LITTTSLLMDD189 pKa = 3.82SEE191 pKa = 4.46SRR193 pKa = 11.84PVQASSTYY201 pKa = 9.99TLVKK205 pKa = 10.24QDD207 pKa = 3.99INTLPLGTPSQLLLQGAVNSYY228 pKa = 10.11EE229 pKa = 3.85NAIAAFGVDD238 pKa = 3.48TSVITYY244 pKa = 9.49SGAMTTQSIEE254 pKa = 4.18DD255 pKa = 3.47VSALNRR261 pKa = 11.84LTMAADD267 pKa = 3.77PATTPVLMPPAMTGLTVADD286 pKa = 4.08ALYY289 pKa = 10.65EE290 pKa = 4.07LGIRR294 pKa = 11.84DD295 pKa = 4.22TNTLAVASTALLANSTLTNVPYY317 pKa = 10.69YY318 pKa = 11.04LEE320 pKa = 4.51FPNFGNCDD328 pKa = 3.55LTEE331 pKa = 4.29LATTSQCDD339 pKa = 3.64AINSHH344 pKa = 5.96WLAAGDD350 pKa = 4.07SPVTILGAVQSGALPPEE367 pKa = 4.48AVLASCPDD375 pKa = 3.67ADD377 pKa = 4.62LSNAASLVGCEE388 pKa = 4.3IKK390 pKa = 10.63DD391 pKa = 3.7AEE393 pKa = 4.49GNPIGLDD400 pKa = 3.3PEE402 pKa = 4.78RR403 pKa = 11.84NLTKK407 pKa = 10.5YY408 pKa = 10.64NPLAAPTEE416 pKa = 4.21MQTLDD421 pKa = 3.68VLVTLPDD428 pKa = 3.3TSDD431 pKa = 3.06AANGVRR437 pKa = 11.84AALGLPAITAAPAAGWPVVVFSHH460 pKa = 7.18GIGGAKK466 pKa = 8.83EE467 pKa = 3.88QNLALAGSFALQGLAMVSIDD487 pKa = 3.88APLHH491 pKa = 5.84GSRR494 pKa = 11.84GWDD497 pKa = 2.78ADD499 pKa = 3.62MNGVFEE505 pKa = 4.87ISASSGMIAFDD516 pKa = 3.42PVKK519 pKa = 10.39YY520 pKa = 10.88ANANVLLYY528 pKa = 11.0VKK530 pKa = 10.37LDD532 pKa = 3.61NLLTTRR538 pKa = 11.84DD539 pKa = 3.53NFRR542 pKa = 11.84QNINDD547 pKa = 3.73HH548 pKa = 6.17LALRR552 pKa = 11.84MGFNSLAQVAPGVFDD567 pKa = 3.58PTRR570 pKa = 11.84VSISGVSLGGINTVSTAAVGGLPLIDD596 pKa = 3.86PTTGSEE602 pKa = 4.19LPSYY606 pKa = 10.24YY607 pKa = 10.72AFSTVMPNVPAQGLAGVFAYY627 pKa = 10.47SEE629 pKa = 4.43TFGPVAKK636 pKa = 10.27AAFVEE641 pKa = 4.35SDD643 pKa = 3.31SFKK646 pKa = 11.0QVLSDD651 pKa = 3.46ATGLSLEE658 pKa = 4.22EE659 pKa = 4.01LAALEE664 pKa = 4.82ASDD667 pKa = 3.97PDD669 pKa = 3.86AYY671 pKa = 10.48QALVDD676 pKa = 3.67QVYY679 pKa = 8.14PTFIAQFVFAAQTVVDD695 pKa = 3.87SSDD698 pKa = 3.44PVGYY702 pKa = 10.77APILAGQGVPVYY714 pKa = 9.94LAEE717 pKa = 4.31VVGDD721 pKa = 3.63TVLPNDD727 pKa = 3.52NSAFGLPLSGTEE739 pKa = 3.61PLIRR743 pKa = 11.84VLGLPGVDD751 pKa = 3.14ATVTGAPATGAVRR764 pKa = 11.84FIDD767 pKa = 3.78SAHH770 pKa = 6.54GSVADD775 pKa = 3.72PTVNPAATVEE785 pKa = 4.31MQTQLAVYY793 pKa = 8.5AASGGAMILVSDD805 pKa = 4.59PSVVQPVMM813 pKa = 3.63
Molecular weight: 83.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A368N695|A0A368N695_9GAMM Erythrose-4-phosphate dehydrogenase OS=Corallincola sp. C4 OX=2282215 GN=DU002_14455 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84SHH16 pKa = 6.15GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38NGRR28 pKa = 11.84ALLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.41GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84SHH16 pKa = 6.15GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38NGRR28 pKa = 11.84ALLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.41GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1271560 |
37 |
11921 |
339.4 |
37.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.595 ± 0.043 | 1.079 ± 0.018 |
5.745 ± 0.039 | 6.162 ± 0.037 |
3.922 ± 0.025 | 7.071 ± 0.035 |
2.253 ± 0.025 | 5.548 ± 0.032 |
4.463 ± 0.038 | 10.934 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.489 ± 0.023 | 3.616 ± 0.029 |
4.265 ± 0.026 | 4.918 ± 0.038 |
5.009 ± 0.037 | 6.412 ± 0.033 |
5.112 ± 0.03 | 7.051 ± 0.045 |
1.362 ± 0.02 | 2.994 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
