
Candida albicans (strain SC5314 / ATCC MYA-2876) (Yeast)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Debaryomycetaceae; Candida/Lodderomyces clade; Candida; Candida albicans
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
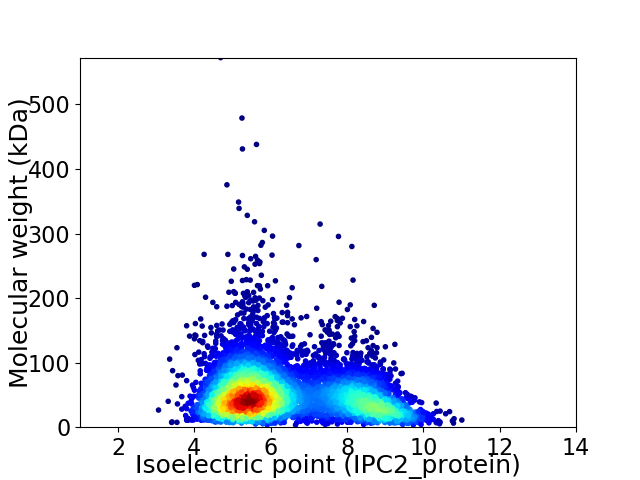
Virtual 2D-PAGE plot for 6036 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
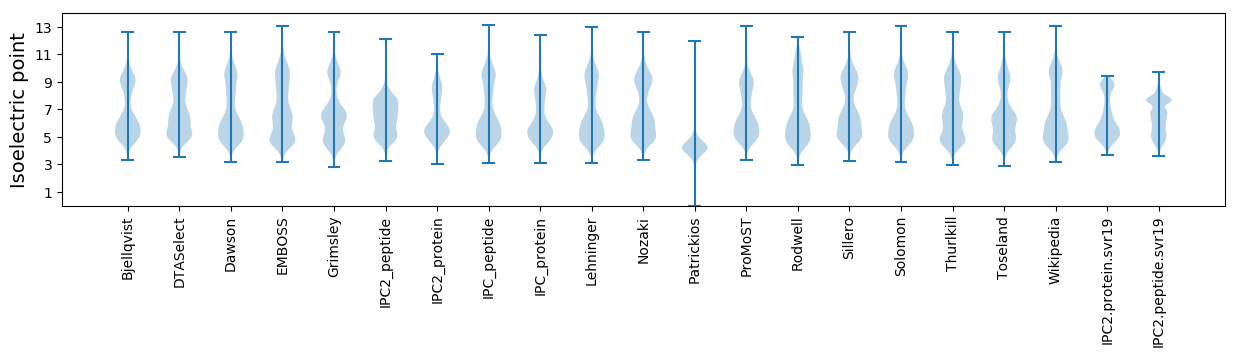
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D8PJY0|A0A1D8PJY0_CANAL DUF2235 domain-containing protein OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=orf19.5894 PE=4 SV=1
MM1 pKa = 7.63DD2 pKa = 5.41HH3 pKa = 7.04SLTSYY8 pKa = 10.72LLSKK12 pKa = 9.51ITTNLEE18 pKa = 3.71CSICSEE24 pKa = 3.9IMIIPMTIEE33 pKa = 4.64CGHH36 pKa = 5.96SFCYY40 pKa = 10.36DD41 pKa = 4.35CIYY44 pKa = 10.9QWFSNKK50 pKa = 9.02INCPTCRR57 pKa = 11.84HH58 pKa = 6.5DD59 pKa = 5.11IEE61 pKa = 4.96NKK63 pKa = 9.79PILNIHH69 pKa = 6.66LKK71 pKa = 9.71EE72 pKa = 3.93ICHH75 pKa = 6.52KK76 pKa = 9.9IIEE79 pKa = 4.59SIIEE83 pKa = 3.97NTSNGDD89 pKa = 3.12GDD91 pKa = 4.3GDD93 pKa = 4.01GDD95 pKa = 4.21SGGVDD100 pKa = 2.66HH101 pKa = 7.39LMNRR105 pKa = 11.84KK106 pKa = 9.27LEE108 pKa = 4.06CLKK111 pKa = 10.24TYY113 pKa = 10.97NLDD116 pKa = 3.68VKK118 pKa = 10.68NKK120 pKa = 10.52SIFGDD125 pKa = 4.04LFNNTVTTLIDD136 pKa = 3.82CSDD139 pKa = 3.21GVARR143 pKa = 11.84CGNCHH148 pKa = 6.0WEE150 pKa = 4.02VHH152 pKa = 5.82GSICSHH158 pKa = 6.62CGTRR162 pKa = 11.84FRR164 pKa = 11.84RR165 pKa = 11.84STRR168 pKa = 11.84NINGEE173 pKa = 4.05SEE175 pKa = 4.02EE176 pKa = 5.32DD177 pKa = 3.45EE178 pKa = 5.82DD179 pKa = 6.24EE180 pKa = 5.29EE181 pKa = 6.97DD182 pKa = 6.23DD183 pKa = 6.53DD184 pKa = 7.44DD185 pKa = 7.73DD186 pKa = 7.61DD187 pKa = 6.83DD188 pKa = 7.32DD189 pKa = 4.85EE190 pKa = 5.87DD191 pKa = 4.32AFEE194 pKa = 4.76DD195 pKa = 4.11VVAGFRR201 pKa = 11.84QSVEE205 pKa = 3.6INEE208 pKa = 4.11YY209 pKa = 11.04DD210 pKa = 3.87SDD212 pKa = 4.62DD213 pKa = 3.99SFIDD217 pKa = 3.27SRR219 pKa = 11.84TANEE223 pKa = 4.25IPVGTDD229 pKa = 2.62SDD231 pKa = 4.54SEE233 pKa = 4.03ISIHH237 pKa = 7.21INANNNNNNNGNVYY251 pKa = 10.78DD252 pKa = 4.27NEE254 pKa = 4.57SNEE257 pKa = 4.19NNNTDD262 pKa = 3.96DD263 pKa = 5.83DD264 pKa = 4.8DD265 pKa = 6.4DD266 pKa = 4.68EE267 pKa = 5.13EE268 pKa = 4.57EE269 pKa = 4.41EE270 pKa = 4.64EE271 pKa = 4.93YY272 pKa = 11.42GDD274 pKa = 3.34NHH276 pKa = 6.98LGHH279 pKa = 6.65TEE281 pKa = 3.96SFGNNSAIHH290 pKa = 6.85DD291 pKa = 4.01DD292 pKa = 3.05WHH294 pKa = 7.73GFEE297 pKa = 5.01SATSSVIDD305 pKa = 3.96RR306 pKa = 11.84AFSDD310 pKa = 3.59DD311 pKa = 4.18DD312 pKa = 3.9EE313 pKa = 5.08EE314 pKa = 4.7EE315 pKa = 4.51EE316 pKa = 4.48EE317 pKa = 4.4DD318 pKa = 4.19VSIQRR323 pKa = 11.84SMNSPYY329 pKa = 11.01NNDD332 pKa = 3.76SIEE335 pKa = 4.23LSSSEE340 pKa = 4.38HH341 pKa = 6.08EE342 pKa = 4.7NIIQLSGDD350 pKa = 4.25DD351 pKa = 4.74EE352 pKa = 6.02DD353 pKa = 5.46DD354 pKa = 3.73SYY356 pKa = 12.14YY357 pKa = 11.03DD358 pKa = 3.94SEE360 pKa = 5.12DD361 pKa = 2.97IRR363 pKa = 11.84DD364 pKa = 3.81AMEE367 pKa = 4.06EE368 pKa = 3.85MDD370 pKa = 5.67NFNIDD375 pKa = 3.21NTDD378 pKa = 3.27YY379 pKa = 11.64SNEE382 pKa = 3.98EE383 pKa = 3.6EE384 pKa = 4.51DD385 pKa = 5.53DD386 pKa = 4.01VVEE389 pKa = 4.2LSDD392 pKa = 3.75YY393 pKa = 11.39NSYY396 pKa = 10.21SDD398 pKa = 5.17NMISDD403 pKa = 3.9VDD405 pKa = 4.15NDD407 pKa = 5.42DD408 pKa = 6.21DD409 pKa = 7.19DD410 pKa = 7.64DD411 pKa = 7.54DD412 pKa = 7.15DD413 pKa = 5.82DD414 pKa = 4.47GWWW417 pKa = 3.15
MM1 pKa = 7.63DD2 pKa = 5.41HH3 pKa = 7.04SLTSYY8 pKa = 10.72LLSKK12 pKa = 9.51ITTNLEE18 pKa = 3.71CSICSEE24 pKa = 3.9IMIIPMTIEE33 pKa = 4.64CGHH36 pKa = 5.96SFCYY40 pKa = 10.36DD41 pKa = 4.35CIYY44 pKa = 10.9QWFSNKK50 pKa = 9.02INCPTCRR57 pKa = 11.84HH58 pKa = 6.5DD59 pKa = 5.11IEE61 pKa = 4.96NKK63 pKa = 9.79PILNIHH69 pKa = 6.66LKK71 pKa = 9.71EE72 pKa = 3.93ICHH75 pKa = 6.52KK76 pKa = 9.9IIEE79 pKa = 4.59SIIEE83 pKa = 3.97NTSNGDD89 pKa = 3.12GDD91 pKa = 4.3GDD93 pKa = 4.01GDD95 pKa = 4.21SGGVDD100 pKa = 2.66HH101 pKa = 7.39LMNRR105 pKa = 11.84KK106 pKa = 9.27LEE108 pKa = 4.06CLKK111 pKa = 10.24TYY113 pKa = 10.97NLDD116 pKa = 3.68VKK118 pKa = 10.68NKK120 pKa = 10.52SIFGDD125 pKa = 4.04LFNNTVTTLIDD136 pKa = 3.82CSDD139 pKa = 3.21GVARR143 pKa = 11.84CGNCHH148 pKa = 6.0WEE150 pKa = 4.02VHH152 pKa = 5.82GSICSHH158 pKa = 6.62CGTRR162 pKa = 11.84FRR164 pKa = 11.84RR165 pKa = 11.84STRR168 pKa = 11.84NINGEE173 pKa = 4.05SEE175 pKa = 4.02EE176 pKa = 5.32DD177 pKa = 3.45EE178 pKa = 5.82DD179 pKa = 6.24EE180 pKa = 5.29EE181 pKa = 6.97DD182 pKa = 6.23DD183 pKa = 6.53DD184 pKa = 7.44DD185 pKa = 7.73DD186 pKa = 7.61DD187 pKa = 6.83DD188 pKa = 7.32DD189 pKa = 4.85EE190 pKa = 5.87DD191 pKa = 4.32AFEE194 pKa = 4.76DD195 pKa = 4.11VVAGFRR201 pKa = 11.84QSVEE205 pKa = 3.6INEE208 pKa = 4.11YY209 pKa = 11.04DD210 pKa = 3.87SDD212 pKa = 4.62DD213 pKa = 3.99SFIDD217 pKa = 3.27SRR219 pKa = 11.84TANEE223 pKa = 4.25IPVGTDD229 pKa = 2.62SDD231 pKa = 4.54SEE233 pKa = 4.03ISIHH237 pKa = 7.21INANNNNNNNGNVYY251 pKa = 10.78DD252 pKa = 4.27NEE254 pKa = 4.57SNEE257 pKa = 4.19NNNTDD262 pKa = 3.96DD263 pKa = 5.83DD264 pKa = 4.8DD265 pKa = 6.4DD266 pKa = 4.68EE267 pKa = 5.13EE268 pKa = 4.57EE269 pKa = 4.41EE270 pKa = 4.64EE271 pKa = 4.93YY272 pKa = 11.42GDD274 pKa = 3.34NHH276 pKa = 6.98LGHH279 pKa = 6.65TEE281 pKa = 3.96SFGNNSAIHH290 pKa = 6.85DD291 pKa = 4.01DD292 pKa = 3.05WHH294 pKa = 7.73GFEE297 pKa = 5.01SATSSVIDD305 pKa = 3.96RR306 pKa = 11.84AFSDD310 pKa = 3.59DD311 pKa = 4.18DD312 pKa = 3.9EE313 pKa = 5.08EE314 pKa = 4.7EE315 pKa = 4.51EE316 pKa = 4.48EE317 pKa = 4.4DD318 pKa = 4.19VSIQRR323 pKa = 11.84SMNSPYY329 pKa = 11.01NNDD332 pKa = 3.76SIEE335 pKa = 4.23LSSSEE340 pKa = 4.38HH341 pKa = 6.08EE342 pKa = 4.7NIIQLSGDD350 pKa = 4.25DD351 pKa = 4.74EE352 pKa = 6.02DD353 pKa = 5.46DD354 pKa = 3.73SYY356 pKa = 12.14YY357 pKa = 11.03DD358 pKa = 3.94SEE360 pKa = 5.12DD361 pKa = 2.97IRR363 pKa = 11.84DD364 pKa = 3.81AMEE367 pKa = 4.06EE368 pKa = 3.85MDD370 pKa = 5.67NFNIDD375 pKa = 3.21NTDD378 pKa = 3.27YY379 pKa = 11.64SNEE382 pKa = 3.98EE383 pKa = 3.6EE384 pKa = 4.51DD385 pKa = 5.53DD386 pKa = 4.01VVEE389 pKa = 4.2LSDD392 pKa = 3.75YY393 pKa = 11.39NSYY396 pKa = 10.21SDD398 pKa = 5.17NMISDD403 pKa = 3.9VDD405 pKa = 4.15NDD407 pKa = 5.42DD408 pKa = 6.21DD409 pKa = 7.19DD410 pKa = 7.64DD411 pKa = 7.54DD412 pKa = 7.15DD413 pKa = 5.82DD414 pKa = 4.47GWWW417 pKa = 3.15
Molecular weight: 47.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D8PN64|A0A1D8PN64_CANAL Aa_trans domain-containing protein OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=orf19.4142 PE=3 SV=1
MM1 pKa = 7.52FSNILRR7 pKa = 11.84QSSRR11 pKa = 11.84GLINFTSVSSRR22 pKa = 11.84PLGVPAFGMNPLITSSPLQSVVGLMQQRR50 pKa = 11.84FKK52 pKa = 11.5SRR54 pKa = 11.84GNTYY58 pKa = 10.2QPSTRR63 pKa = 11.84KK64 pKa = 9.59RR65 pKa = 11.84KK66 pKa = 9.61RR67 pKa = 11.84KK68 pKa = 9.79LGFLARR74 pKa = 11.84LRR76 pKa = 11.84SIGGRR81 pKa = 11.84KK82 pKa = 8.48ILEE85 pKa = 3.7RR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84AKK90 pKa = 10.52GRR92 pKa = 11.84WFLSHH97 pKa = 7.05
MM1 pKa = 7.52FSNILRR7 pKa = 11.84QSSRR11 pKa = 11.84GLINFTSVSSRR22 pKa = 11.84PLGVPAFGMNPLITSSPLQSVVGLMQQRR50 pKa = 11.84FKK52 pKa = 11.5SRR54 pKa = 11.84GNTYY58 pKa = 10.2QPSTRR63 pKa = 11.84KK64 pKa = 9.59RR65 pKa = 11.84KK66 pKa = 9.61RR67 pKa = 11.84KK68 pKa = 9.79LGFLARR74 pKa = 11.84LRR76 pKa = 11.84SIGGRR81 pKa = 11.84KK82 pKa = 8.48ILEE85 pKa = 3.7RR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84AKK90 pKa = 10.52GRR92 pKa = 11.84WFLSHH97 pKa = 7.05
Molecular weight: 11.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2982981 |
31 |
5035 |
494.2 |
55.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.049 ± 0.024 | 1.067 ± 0.01 |
5.905 ± 0.025 | 6.467 ± 0.033 |
4.396 ± 0.02 | 5.079 ± 0.025 |
2.09 ± 0.012 | 7.114 ± 0.027 |
7.348 ± 0.033 | 9.209 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.782 ± 0.009 | 6.691 ± 0.032 |
4.473 ± 0.024 | 4.493 ± 0.031 |
3.763 ± 0.021 | 8.976 ± 0.046 |
6.146 ± 0.031 | 5.429 ± 0.024 |
0.969 ± 0.01 | 3.55 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
