
Meiothermus phage MMP7
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; unclassified Myoviridae
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
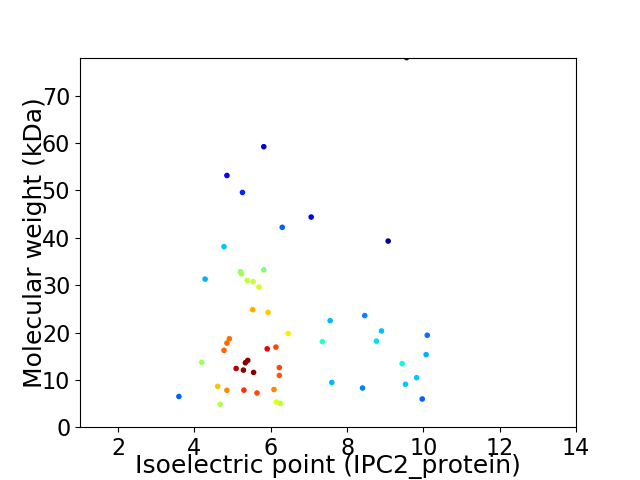
Virtual 2D-PAGE plot for 53 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G8F4V1|A0A3G8F4V1_9CAUD Uncharacterized protein OS=Meiothermus phage MMP7 OX=2483850 PE=4 SV=1
MM1 pKa = 7.24CQEE4 pKa = 4.39CQGCGYY10 pKa = 10.11VEE12 pKa = 4.32YY13 pKa = 10.92VAGFYY18 pKa = 10.83FSDD21 pKa = 3.39SFGNYY26 pKa = 9.42LPRR29 pKa = 11.84EE30 pKa = 4.13EE31 pKa = 4.13IARR34 pKa = 11.84CEE36 pKa = 4.28VCDD39 pKa = 3.43GTGFVEE45 pKa = 5.14VWDD48 pKa = 3.73EE49 pKa = 4.3DD50 pKa = 4.06EE51 pKa = 5.73AVLEE55 pKa = 4.42GDD57 pKa = 3.85GG58 pKa = 4.34
MM1 pKa = 7.24CQEE4 pKa = 4.39CQGCGYY10 pKa = 10.11VEE12 pKa = 4.32YY13 pKa = 10.92VAGFYY18 pKa = 10.83FSDD21 pKa = 3.39SFGNYY26 pKa = 9.42LPRR29 pKa = 11.84EE30 pKa = 4.13EE31 pKa = 4.13IARR34 pKa = 11.84CEE36 pKa = 4.28VCDD39 pKa = 3.43GTGFVEE45 pKa = 5.14VWDD48 pKa = 3.73EE49 pKa = 4.3DD50 pKa = 4.06EE51 pKa = 5.73AVLEE55 pKa = 4.42GDD57 pKa = 3.85GG58 pKa = 4.34
Molecular weight: 6.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G8F535|A0A3G8F535_9CAUD Major capsid protein OS=Meiothermus phage MMP7 OX=2483850 PE=4 SV=1
MM1 pKa = 7.53TRR3 pKa = 11.84LQWTFDD9 pKa = 3.5LLDD12 pKa = 4.01RR13 pKa = 11.84VSAPARR19 pKa = 11.84RR20 pKa = 11.84IGQALKK26 pKa = 9.38NTDD29 pKa = 3.25AALKK33 pKa = 9.68KK34 pKa = 10.22VAGGGEE40 pKa = 4.05AAQKK44 pKa = 10.56ALQRR48 pKa = 11.84SFGLSEE54 pKa = 3.76RR55 pKa = 11.84AAVRR59 pKa = 11.84LTTAIATVGQVQGAIGGLQARR80 pKa = 11.84VGALRR85 pKa = 11.84GAFSSVTDD93 pKa = 3.6SVFNLKK99 pKa = 10.01NALVAGAIGFATKK112 pKa = 10.48VVIDD116 pKa = 3.36QVGFIEE122 pKa = 4.15QQRR125 pKa = 11.84IALGTILGSPIRR137 pKa = 11.84ATAALDD143 pKa = 3.47WAIKK147 pKa = 9.8FADD150 pKa = 3.61VTPFEE155 pKa = 4.44TPQVLEE161 pKa = 4.31AMRR164 pKa = 11.84QALAVGFNTKK174 pKa = 10.08QIEE177 pKa = 4.18PLLTTLGDD185 pKa = 3.45TAAALSLGPAGLNDD199 pKa = 5.62LITVFGQIRR208 pKa = 11.84SAGKK212 pKa = 10.54LNTEE216 pKa = 4.37EE217 pKa = 4.4VNQLTEE223 pKa = 3.94RR224 pKa = 11.84GIPAFEE230 pKa = 4.06ILAKK234 pKa = 10.57AFSTDD239 pKa = 2.85IPTVRR244 pKa = 11.84QMIEE248 pKa = 3.48KK249 pKa = 10.12GQIASEE255 pKa = 4.05AALGAIYY262 pKa = 10.33RR263 pKa = 11.84GLKK266 pKa = 8.28TRR268 pKa = 11.84FGGGMAAQSRR278 pKa = 11.84SIFGLVSTLRR288 pKa = 11.84SRR290 pKa = 11.84PQTIAFRR297 pKa = 11.84LEE299 pKa = 3.49RR300 pKa = 11.84DD301 pKa = 3.25KK302 pKa = 11.51ALEE305 pKa = 3.81PFRR308 pKa = 11.84RR309 pKa = 11.84LLMNLADD316 pKa = 3.75LTDD319 pKa = 4.1FNKK322 pKa = 10.55PPGSTIAARR331 pKa = 11.84LTSGIGGLFKK341 pKa = 10.8AAFGPLAAATEE352 pKa = 4.2PKK354 pKa = 10.24RR355 pKa = 11.84AGEE358 pKa = 3.89AVLAFINRR366 pKa = 11.84ATAAVNRR373 pKa = 11.84ARR375 pKa = 11.84DD376 pKa = 3.17AWPAVKK382 pKa = 9.96AAALDD387 pKa = 4.42FISGVRR393 pKa = 11.84SGFDD397 pKa = 3.25LLAGIWRR404 pKa = 11.84TVEE407 pKa = 3.88PLVSGLGRR415 pKa = 11.84LAGSFTGAQASMGGANINAVKK436 pKa = 10.44IIGTIAALAAVWRR449 pKa = 11.84VLNLVTLGGAGAIARR464 pKa = 11.84WGAIAVVSLVRR475 pKa = 11.84VAAVGLPILWAKK487 pKa = 10.41IGALNTLGVTALRR500 pKa = 11.84ASGRR504 pKa = 11.84ALLAGGRR511 pKa = 11.84MALAWIIGLGPIGWLIGGVAAISGALVLAYY541 pKa = 10.78NKK543 pKa = 9.87VDD545 pKa = 3.09WFRR548 pKa = 11.84NVVNRR553 pKa = 11.84AWEE556 pKa = 4.29SIKK559 pKa = 10.59QVGKK563 pKa = 10.95GLLDD567 pKa = 3.5WFTSLPTRR575 pKa = 11.84IGQTFAQLPNLLRR588 pKa = 11.84GLLRR592 pKa = 11.84RR593 pKa = 11.84AIDD596 pKa = 3.82LLPPGVRR603 pKa = 11.84DD604 pKa = 3.65VVRR607 pKa = 11.84GLVGGLLDD615 pKa = 4.8GSDD618 pKa = 3.78PVEE621 pKa = 4.22NAATQLADD629 pKa = 3.22KK630 pKa = 8.07TQQGFARR637 pKa = 11.84PLEE640 pKa = 4.0IRR642 pKa = 11.84SPSRR646 pKa = 11.84RR647 pKa = 11.84FAYY650 pKa = 9.48YY651 pKa = 10.07GQMMGAGLEE660 pKa = 4.0VGMRR664 pKa = 11.84DD665 pKa = 2.93SLGRR669 pKa = 11.84VQRR672 pKa = 11.84AAAGMTIAATLAMSGGPALSVAPPALAPTTRR703 pKa = 11.84AISITVGPITVNGGSDD719 pKa = 3.24TKK721 pKa = 11.0QIADD725 pKa = 3.92EE726 pKa = 4.25LRR728 pKa = 11.84TVAVEE733 pKa = 4.15AVLEE737 pKa = 4.13ALEE740 pKa = 4.32RR741 pKa = 11.84AAHH744 pKa = 6.06EE745 pKa = 4.44EE746 pKa = 4.36GAA748 pKa = 3.95
MM1 pKa = 7.53TRR3 pKa = 11.84LQWTFDD9 pKa = 3.5LLDD12 pKa = 4.01RR13 pKa = 11.84VSAPARR19 pKa = 11.84RR20 pKa = 11.84IGQALKK26 pKa = 9.38NTDD29 pKa = 3.25AALKK33 pKa = 9.68KK34 pKa = 10.22VAGGGEE40 pKa = 4.05AAQKK44 pKa = 10.56ALQRR48 pKa = 11.84SFGLSEE54 pKa = 3.76RR55 pKa = 11.84AAVRR59 pKa = 11.84LTTAIATVGQVQGAIGGLQARR80 pKa = 11.84VGALRR85 pKa = 11.84GAFSSVTDD93 pKa = 3.6SVFNLKK99 pKa = 10.01NALVAGAIGFATKK112 pKa = 10.48VVIDD116 pKa = 3.36QVGFIEE122 pKa = 4.15QQRR125 pKa = 11.84IALGTILGSPIRR137 pKa = 11.84ATAALDD143 pKa = 3.47WAIKK147 pKa = 9.8FADD150 pKa = 3.61VTPFEE155 pKa = 4.44TPQVLEE161 pKa = 4.31AMRR164 pKa = 11.84QALAVGFNTKK174 pKa = 10.08QIEE177 pKa = 4.18PLLTTLGDD185 pKa = 3.45TAAALSLGPAGLNDD199 pKa = 5.62LITVFGQIRR208 pKa = 11.84SAGKK212 pKa = 10.54LNTEE216 pKa = 4.37EE217 pKa = 4.4VNQLTEE223 pKa = 3.94RR224 pKa = 11.84GIPAFEE230 pKa = 4.06ILAKK234 pKa = 10.57AFSTDD239 pKa = 2.85IPTVRR244 pKa = 11.84QMIEE248 pKa = 3.48KK249 pKa = 10.12GQIASEE255 pKa = 4.05AALGAIYY262 pKa = 10.33RR263 pKa = 11.84GLKK266 pKa = 8.28TRR268 pKa = 11.84FGGGMAAQSRR278 pKa = 11.84SIFGLVSTLRR288 pKa = 11.84SRR290 pKa = 11.84PQTIAFRR297 pKa = 11.84LEE299 pKa = 3.49RR300 pKa = 11.84DD301 pKa = 3.25KK302 pKa = 11.51ALEE305 pKa = 3.81PFRR308 pKa = 11.84RR309 pKa = 11.84LLMNLADD316 pKa = 3.75LTDD319 pKa = 4.1FNKK322 pKa = 10.55PPGSTIAARR331 pKa = 11.84LTSGIGGLFKK341 pKa = 10.8AAFGPLAAATEE352 pKa = 4.2PKK354 pKa = 10.24RR355 pKa = 11.84AGEE358 pKa = 3.89AVLAFINRR366 pKa = 11.84ATAAVNRR373 pKa = 11.84ARR375 pKa = 11.84DD376 pKa = 3.17AWPAVKK382 pKa = 9.96AAALDD387 pKa = 4.42FISGVRR393 pKa = 11.84SGFDD397 pKa = 3.25LLAGIWRR404 pKa = 11.84TVEE407 pKa = 3.88PLVSGLGRR415 pKa = 11.84LAGSFTGAQASMGGANINAVKK436 pKa = 10.44IIGTIAALAAVWRR449 pKa = 11.84VLNLVTLGGAGAIARR464 pKa = 11.84WGAIAVVSLVRR475 pKa = 11.84VAAVGLPILWAKK487 pKa = 10.41IGALNTLGVTALRR500 pKa = 11.84ASGRR504 pKa = 11.84ALLAGGRR511 pKa = 11.84MALAWIIGLGPIGWLIGGVAAISGALVLAYY541 pKa = 10.78NKK543 pKa = 9.87VDD545 pKa = 3.09WFRR548 pKa = 11.84NVVNRR553 pKa = 11.84AWEE556 pKa = 4.29SIKK559 pKa = 10.59QVGKK563 pKa = 10.95GLLDD567 pKa = 3.5WFTSLPTRR575 pKa = 11.84IGQTFAQLPNLLRR588 pKa = 11.84GLLRR592 pKa = 11.84RR593 pKa = 11.84AIDD596 pKa = 3.82LLPPGVRR603 pKa = 11.84DD604 pKa = 3.65VVRR607 pKa = 11.84GLVGGLLDD615 pKa = 4.8GSDD618 pKa = 3.78PVEE621 pKa = 4.22NAATQLADD629 pKa = 3.22KK630 pKa = 8.07TQQGFARR637 pKa = 11.84PLEE640 pKa = 4.0IRR642 pKa = 11.84SPSRR646 pKa = 11.84RR647 pKa = 11.84FAYY650 pKa = 9.48YY651 pKa = 10.07GQMMGAGLEE660 pKa = 4.0VGMRR664 pKa = 11.84DD665 pKa = 2.93SLGRR669 pKa = 11.84VQRR672 pKa = 11.84AAAGMTIAATLAMSGGPALSVAPPALAPTTRR703 pKa = 11.84AISITVGPITVNGGSDD719 pKa = 3.24TKK721 pKa = 11.0QIADD725 pKa = 3.92EE726 pKa = 4.25LRR728 pKa = 11.84TVAVEE733 pKa = 4.15AVLEE737 pKa = 4.13ALEE740 pKa = 4.32RR741 pKa = 11.84AAHH744 pKa = 6.06EE745 pKa = 4.44EE746 pKa = 4.36GAA748 pKa = 3.95
Molecular weight: 78.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10268 |
40 |
748 |
193.7 |
21.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.096 ± 0.514 | 0.779 ± 0.167 |
4.704 ± 0.213 | 7.372 ± 0.586 |
2.376 ± 0.201 | 8.239 ± 0.408 |
1.529 ± 0.223 | 4.1 ± 0.265 |
2.649 ± 0.276 | 11.102 ± 0.37 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.558 ± 0.095 | 2.678 ± 0.224 |
6.048 ± 0.347 | 3.759 ± 0.192 |
8.668 ± 0.432 | 4.48 ± 0.159 |
5.619 ± 0.37 | 6.515 ± 0.311 |
2.707 ± 0.208 | 3.019 ± 0.246 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
