
Blastococcus sp. CCUG 61487
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Blastococcus; unclassified Blastococcus
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
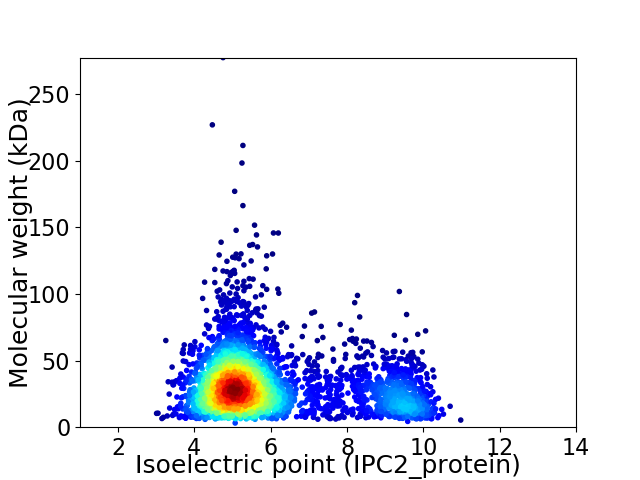
Virtual 2D-PAGE plot for 3688 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V5TZJ7|A0A4V5TZJ7_9ACTN Pilus assembly protein CpaF OS=Blastococcus sp. CCUG 61487 OX=1840703 GN=A6V29_08360 PE=3 SV=1
MM1 pKa = 7.6AGAGGRR7 pKa = 11.84RR8 pKa = 11.84AALLPAVLACLLVLTGCSMVVVGQAIPALPPVTDD42 pKa = 4.23AEE44 pKa = 4.36PGEE47 pKa = 4.22IEE49 pKa = 4.6VVGATDD55 pKa = 3.71QEE57 pKa = 4.4VDD59 pKa = 3.52VLARR63 pKa = 11.84NALADD68 pKa = 4.85LEE70 pKa = 4.8TFWAEE75 pKa = 3.48QQPAVFGLPFEE86 pKa = 4.53PLEE89 pKa = 4.32GGYY92 pKa = 10.63FSVDD96 pKa = 3.21PGDD99 pKa = 3.48VDD101 pKa = 3.45PRR103 pKa = 11.84LYY105 pKa = 9.23PTGIGCGADD114 pKa = 3.3PRR116 pKa = 11.84EE117 pKa = 4.28VEE119 pKa = 4.31DD120 pKa = 3.43NAYY123 pKa = 10.42YY124 pKa = 10.75CLARR128 pKa = 11.84EE129 pKa = 4.72APNSDD134 pKa = 3.81AISYY138 pKa = 10.59DD139 pKa = 3.25RR140 pKa = 11.84TFLGDD145 pKa = 3.81LGADD149 pKa = 4.19FGRR152 pKa = 11.84FIPALVMAHH161 pKa = 6.2EE162 pKa = 5.31FGHH165 pKa = 6.69AVQARR170 pKa = 11.84VGSPEE175 pKa = 3.63FSILVEE181 pKa = 4.2TQADD185 pKa = 4.05CLAGAWSRR193 pKa = 11.84WVADD197 pKa = 3.71GAAEE201 pKa = 4.1HH202 pKa = 5.91TVLRR206 pKa = 11.84TSEE209 pKa = 4.27LDD211 pKa = 3.47DD212 pKa = 3.59VLRR215 pKa = 11.84GYY217 pKa = 11.18LLLRR221 pKa = 11.84DD222 pKa = 3.96PVGTNPGQPQAHH234 pKa = 6.42GSYY237 pKa = 10.16FDD239 pKa = 3.68RR240 pKa = 11.84VSAFQEE246 pKa = 4.44GFEE249 pKa = 4.82LGAEE253 pKa = 4.16ACRR256 pKa = 11.84DD257 pKa = 3.69NFDD260 pKa = 3.19VARR263 pKa = 11.84PLTQDD268 pKa = 3.06RR269 pKa = 11.84FRR271 pKa = 11.84TVGEE275 pKa = 4.03IEE277 pKa = 4.2TGGNAPYY284 pKa = 10.46EE285 pKa = 4.23VLLDD289 pKa = 3.79IAAEE293 pKa = 4.1TLPQAWASALGDD305 pKa = 3.77DD306 pKa = 4.6FRR308 pKa = 11.84PPSVEE313 pKa = 3.96RR314 pKa = 11.84FDD316 pKa = 5.84GEE318 pKa = 4.93APDD321 pKa = 4.52CAPDD325 pKa = 4.19PGLDD329 pKa = 4.34LVYY332 pKa = 10.8CPDD335 pKa = 4.81DD336 pKa = 4.0GLVAFDD342 pKa = 4.11EE343 pKa = 4.56TDD345 pKa = 3.48LAVPAYY351 pKa = 10.13EE352 pKa = 4.11EE353 pKa = 4.66LGDD356 pKa = 3.88FAVATAISLPYY367 pKa = 9.96GLAVRR372 pKa = 11.84DD373 pKa = 3.79QLGLSVDD380 pKa = 4.27DD381 pKa = 4.04EE382 pKa = 4.45DD383 pKa = 3.99AARR386 pKa = 11.84SVVCLSGWYY395 pKa = 10.02AEE397 pKa = 4.3QLYY400 pKa = 10.37SDD402 pKa = 4.61PLGADD407 pKa = 4.44GIFTSSISPGDD418 pKa = 3.26VDD420 pKa = 4.71EE421 pKa = 5.08SVQFLLTFGSEE432 pKa = 4.33PEE434 pKa = 4.14VLPGVDD440 pKa = 3.24LTGFQLVDD448 pKa = 4.03LYY450 pKa = 11.14RR451 pKa = 11.84AGFFEE456 pKa = 5.45GLDD459 pKa = 3.5ACGLL463 pKa = 3.81
MM1 pKa = 7.6AGAGGRR7 pKa = 11.84RR8 pKa = 11.84AALLPAVLACLLVLTGCSMVVVGQAIPALPPVTDD42 pKa = 4.23AEE44 pKa = 4.36PGEE47 pKa = 4.22IEE49 pKa = 4.6VVGATDD55 pKa = 3.71QEE57 pKa = 4.4VDD59 pKa = 3.52VLARR63 pKa = 11.84NALADD68 pKa = 4.85LEE70 pKa = 4.8TFWAEE75 pKa = 3.48QQPAVFGLPFEE86 pKa = 4.53PLEE89 pKa = 4.32GGYY92 pKa = 10.63FSVDD96 pKa = 3.21PGDD99 pKa = 3.48VDD101 pKa = 3.45PRR103 pKa = 11.84LYY105 pKa = 9.23PTGIGCGADD114 pKa = 3.3PRR116 pKa = 11.84EE117 pKa = 4.28VEE119 pKa = 4.31DD120 pKa = 3.43NAYY123 pKa = 10.42YY124 pKa = 10.75CLARR128 pKa = 11.84EE129 pKa = 4.72APNSDD134 pKa = 3.81AISYY138 pKa = 10.59DD139 pKa = 3.25RR140 pKa = 11.84TFLGDD145 pKa = 3.81LGADD149 pKa = 4.19FGRR152 pKa = 11.84FIPALVMAHH161 pKa = 6.2EE162 pKa = 5.31FGHH165 pKa = 6.69AVQARR170 pKa = 11.84VGSPEE175 pKa = 3.63FSILVEE181 pKa = 4.2TQADD185 pKa = 4.05CLAGAWSRR193 pKa = 11.84WVADD197 pKa = 3.71GAAEE201 pKa = 4.1HH202 pKa = 5.91TVLRR206 pKa = 11.84TSEE209 pKa = 4.27LDD211 pKa = 3.47DD212 pKa = 3.59VLRR215 pKa = 11.84GYY217 pKa = 11.18LLLRR221 pKa = 11.84DD222 pKa = 3.96PVGTNPGQPQAHH234 pKa = 6.42GSYY237 pKa = 10.16FDD239 pKa = 3.68RR240 pKa = 11.84VSAFQEE246 pKa = 4.44GFEE249 pKa = 4.82LGAEE253 pKa = 4.16ACRR256 pKa = 11.84DD257 pKa = 3.69NFDD260 pKa = 3.19VARR263 pKa = 11.84PLTQDD268 pKa = 3.06RR269 pKa = 11.84FRR271 pKa = 11.84TVGEE275 pKa = 4.03IEE277 pKa = 4.2TGGNAPYY284 pKa = 10.46EE285 pKa = 4.23VLLDD289 pKa = 3.79IAAEE293 pKa = 4.1TLPQAWASALGDD305 pKa = 3.77DD306 pKa = 4.6FRR308 pKa = 11.84PPSVEE313 pKa = 3.96RR314 pKa = 11.84FDD316 pKa = 5.84GEE318 pKa = 4.93APDD321 pKa = 4.52CAPDD325 pKa = 4.19PGLDD329 pKa = 4.34LVYY332 pKa = 10.8CPDD335 pKa = 4.81DD336 pKa = 4.0GLVAFDD342 pKa = 4.11EE343 pKa = 4.56TDD345 pKa = 3.48LAVPAYY351 pKa = 10.13EE352 pKa = 4.11EE353 pKa = 4.66LGDD356 pKa = 3.88FAVATAISLPYY367 pKa = 9.96GLAVRR372 pKa = 11.84DD373 pKa = 3.79QLGLSVDD380 pKa = 4.27DD381 pKa = 4.04EE382 pKa = 4.45DD383 pKa = 3.99AARR386 pKa = 11.84SVVCLSGWYY395 pKa = 10.02AEE397 pKa = 4.3QLYY400 pKa = 10.37SDD402 pKa = 4.61PLGADD407 pKa = 4.44GIFTSSISPGDD418 pKa = 3.26VDD420 pKa = 4.71EE421 pKa = 5.08SVQFLLTFGSEE432 pKa = 4.33PEE434 pKa = 4.14VLPGVDD440 pKa = 3.24LTGFQLVDD448 pKa = 4.03LYY450 pKa = 11.14RR451 pKa = 11.84AGFFEE456 pKa = 5.45GLDD459 pKa = 3.5ACGLL463 pKa = 3.81
Molecular weight: 49.26 kDa
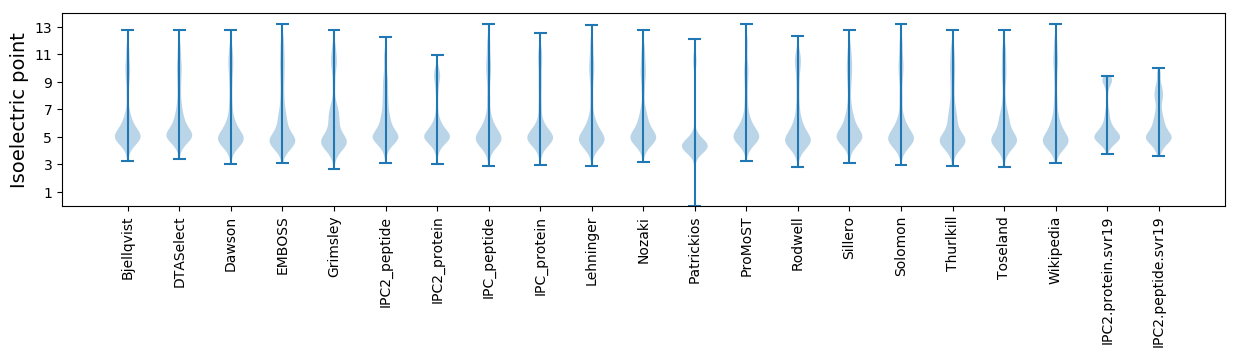
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V5TZR4|A0A4V5TZR4_9ACTN Hydrolase OS=Blastococcus sp. CCUG 61487 OX=1840703 GN=A6V29_07305 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPNTRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 8.65THH17 pKa = 5.48GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAGRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 10.08GRR40 pKa = 11.84EE41 pKa = 3.61KK42 pKa = 10.93LSAA45 pKa = 3.78
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPNTRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 8.65THH17 pKa = 5.48GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAGRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 10.08GRR40 pKa = 11.84EE41 pKa = 3.61KK42 pKa = 10.93LSAA45 pKa = 3.78
Molecular weight: 5.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1142701 |
29 |
2637 |
309.8 |
33.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.992 ± 0.064 | 0.686 ± 0.011 |
6.392 ± 0.03 | 5.92 ± 0.033 |
2.587 ± 0.024 | 9.372 ± 0.041 |
2.014 ± 0.017 | 3.055 ± 0.029 |
1.519 ± 0.029 | 10.69 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.667 ± 0.015 | 1.496 ± 0.02 |
6.086 ± 0.033 | 2.764 ± 0.022 |
8.214 ± 0.043 | 4.836 ± 0.029 |
5.882 ± 0.033 | 9.526 ± 0.041 |
1.511 ± 0.017 | 1.791 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
