
Desulfococcus oleovorans (strain DSM 6200 / JCM 39069 / Hxd3)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobacteraceae; Desulfococcus; Desulfococcus oleovorans
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
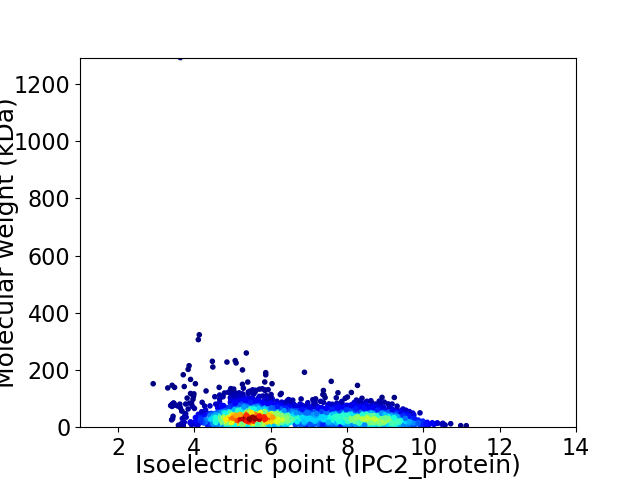
Virtual 2D-PAGE plot for 3255 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
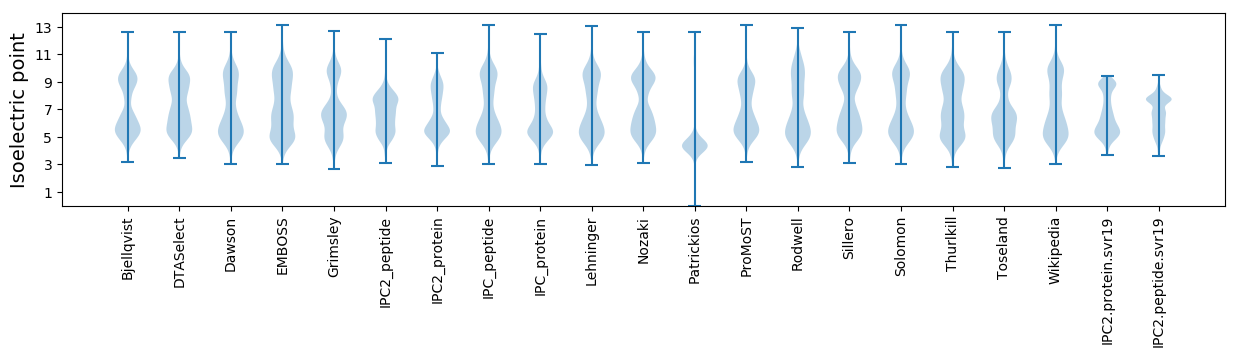
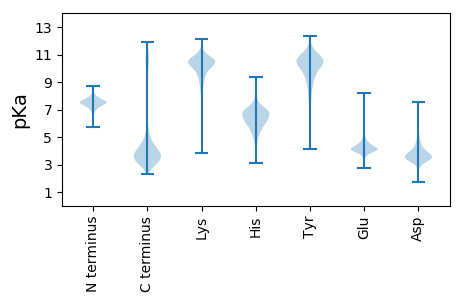
Protein with the lowest isoelectric point:
>tr|A9A0Q7|A9A0Q7_DESOH Response regulator receiver protein OS=Desulfococcus oleovorans (strain DSM 6200 / JCM 39069 / Hxd3) OX=96561 GN=Dole_3271 PE=4 SV=1
MM1 pKa = 7.62PMNYY5 pKa = 9.49HH6 pKa = 6.83KK7 pKa = 10.21KK8 pKa = 6.45TTAGVRR14 pKa = 11.84NRR16 pKa = 11.84ILWAGLIPLVLCALFVLPAGATDD39 pKa = 3.51IEE41 pKa = 4.55TTPRR45 pKa = 11.84YY46 pKa = 8.41TFVGNVNFVGTGGSLRR62 pKa = 11.84TRR64 pKa = 11.84PNAGWWADD72 pKa = 3.41PCRR75 pKa = 11.84ITTTSTEE82 pKa = 4.09ALSGIPAGATIRR94 pKa = 11.84AAYY97 pKa = 9.5LYY99 pKa = 8.32WARR102 pKa = 11.84SGNAADD108 pKa = 4.15NQVSLNGNTVNADD121 pKa = 3.63RR122 pKa = 11.84PFDD125 pKa = 3.55EE126 pKa = 4.59TFVNGGTTYY135 pKa = 11.13RR136 pKa = 11.84FFSGMADD143 pKa = 3.13VTSIVNTTRR152 pKa = 11.84NGNYY156 pKa = 9.1TFGGLTISAGNPWCGVQAVVGGWALVVIYY185 pKa = 10.23EE186 pKa = 4.26DD187 pKa = 3.56ASEE190 pKa = 4.25KK191 pKa = 10.58EE192 pKa = 4.16RR193 pKa = 11.84VINIYY198 pKa = 10.87DD199 pKa = 3.96GFQYY203 pKa = 10.53FRR205 pKa = 11.84DD206 pKa = 3.6SDD208 pKa = 3.87IVLTPSNFVVSSASGIDD225 pKa = 3.38GKK227 pKa = 10.47IGHH230 pKa = 6.98ISWEE234 pKa = 4.18GDD236 pKa = 3.29PTLTGANEE244 pKa = 3.91YY245 pKa = 10.86LRR247 pKa = 11.84VNGTTLTDD255 pKa = 3.75ASNSATDD262 pKa = 3.58QYY264 pKa = 11.74NSTFSHH270 pKa = 7.12LGTLVSSPTYY280 pKa = 10.62GVDD283 pKa = 3.61FDD285 pKa = 4.76IYY287 pKa = 10.76DD288 pKa = 3.53ISSLLSEE295 pKa = 4.86GDD297 pKa = 3.53SSLSSVYY304 pKa = 10.41SAAGDD309 pKa = 3.78LVILSAEE316 pKa = 4.44IISVTCQAADD326 pKa = 3.57LSISKK331 pKa = 7.54THH333 pKa = 6.12SGGFVSGGTGDD344 pKa = 3.54FTITVTNSGPNSEE357 pKa = 4.51TGTVTVTDD365 pKa = 3.85VLPAGLTYY373 pKa = 10.9SSFSGTGWAVDD384 pKa = 3.42TSGAPTIVFTYY395 pKa = 10.14DD396 pKa = 3.33CSASPLPPGNSLPALTLTVNVGAAAVPNVSNTASVDD432 pKa = 3.13SDD434 pKa = 3.75TTFDD438 pKa = 5.14PDD440 pKa = 3.45TSDD443 pKa = 4.68NSDD446 pKa = 3.33TDD448 pKa = 3.84TVTVSAAAPDD458 pKa = 4.2LLVMKK463 pKa = 9.36TVTTFSDD470 pKa = 3.62PTGGANPKK478 pKa = 9.65AIPGAVMIYY487 pKa = 9.21TIQVTNQGAGPVDD500 pKa = 3.68ADD502 pKa = 4.51SINITDD508 pKa = 6.15AIPANTKK515 pKa = 10.61LFVGDD520 pKa = 4.77LGDD523 pKa = 3.96GPIVFLDD530 pKa = 4.23GSAYY534 pKa = 10.39GGTDD538 pKa = 3.18SGLDD542 pKa = 3.47PYY544 pKa = 11.58DD545 pKa = 4.83YY546 pKa = 10.76IALDD550 pKa = 3.98NDD552 pKa = 3.7TDD554 pKa = 4.16DD555 pKa = 5.96LGFSSDD561 pKa = 3.26NGATFTYY568 pKa = 10.27HH569 pKa = 6.23PTPDD573 pKa = 4.87GDD575 pKa = 4.35DD576 pKa = 3.54CDD578 pKa = 4.95EE579 pKa = 4.34NVTDD583 pKa = 4.86ILVNPKK589 pKa = 10.74GPMDD593 pKa = 4.36GAASGNTPSFQVRR606 pKa = 11.84FRR608 pKa = 11.84VQVEE612 pKa = 3.79
MM1 pKa = 7.62PMNYY5 pKa = 9.49HH6 pKa = 6.83KK7 pKa = 10.21KK8 pKa = 6.45TTAGVRR14 pKa = 11.84NRR16 pKa = 11.84ILWAGLIPLVLCALFVLPAGATDD39 pKa = 3.51IEE41 pKa = 4.55TTPRR45 pKa = 11.84YY46 pKa = 8.41TFVGNVNFVGTGGSLRR62 pKa = 11.84TRR64 pKa = 11.84PNAGWWADD72 pKa = 3.41PCRR75 pKa = 11.84ITTTSTEE82 pKa = 4.09ALSGIPAGATIRR94 pKa = 11.84AAYY97 pKa = 9.5LYY99 pKa = 8.32WARR102 pKa = 11.84SGNAADD108 pKa = 4.15NQVSLNGNTVNADD121 pKa = 3.63RR122 pKa = 11.84PFDD125 pKa = 3.55EE126 pKa = 4.59TFVNGGTTYY135 pKa = 11.13RR136 pKa = 11.84FFSGMADD143 pKa = 3.13VTSIVNTTRR152 pKa = 11.84NGNYY156 pKa = 9.1TFGGLTISAGNPWCGVQAVVGGWALVVIYY185 pKa = 10.23EE186 pKa = 4.26DD187 pKa = 3.56ASEE190 pKa = 4.25KK191 pKa = 10.58EE192 pKa = 4.16RR193 pKa = 11.84VINIYY198 pKa = 10.87DD199 pKa = 3.96GFQYY203 pKa = 10.53FRR205 pKa = 11.84DD206 pKa = 3.6SDD208 pKa = 3.87IVLTPSNFVVSSASGIDD225 pKa = 3.38GKK227 pKa = 10.47IGHH230 pKa = 6.98ISWEE234 pKa = 4.18GDD236 pKa = 3.29PTLTGANEE244 pKa = 3.91YY245 pKa = 10.86LRR247 pKa = 11.84VNGTTLTDD255 pKa = 3.75ASNSATDD262 pKa = 3.58QYY264 pKa = 11.74NSTFSHH270 pKa = 7.12LGTLVSSPTYY280 pKa = 10.62GVDD283 pKa = 3.61FDD285 pKa = 4.76IYY287 pKa = 10.76DD288 pKa = 3.53ISSLLSEE295 pKa = 4.86GDD297 pKa = 3.53SSLSSVYY304 pKa = 10.41SAAGDD309 pKa = 3.78LVILSAEE316 pKa = 4.44IISVTCQAADD326 pKa = 3.57LSISKK331 pKa = 7.54THH333 pKa = 6.12SGGFVSGGTGDD344 pKa = 3.54FTITVTNSGPNSEE357 pKa = 4.51TGTVTVTDD365 pKa = 3.85VLPAGLTYY373 pKa = 10.9SSFSGTGWAVDD384 pKa = 3.42TSGAPTIVFTYY395 pKa = 10.14DD396 pKa = 3.33CSASPLPPGNSLPALTLTVNVGAAAVPNVSNTASVDD432 pKa = 3.13SDD434 pKa = 3.75TTFDD438 pKa = 5.14PDD440 pKa = 3.45TSDD443 pKa = 4.68NSDD446 pKa = 3.33TDD448 pKa = 3.84TVTVSAAAPDD458 pKa = 4.2LLVMKK463 pKa = 9.36TVTTFSDD470 pKa = 3.62PTGGANPKK478 pKa = 9.65AIPGAVMIYY487 pKa = 9.21TIQVTNQGAGPVDD500 pKa = 3.68ADD502 pKa = 4.51SINITDD508 pKa = 6.15AIPANTKK515 pKa = 10.61LFVGDD520 pKa = 4.77LGDD523 pKa = 3.96GPIVFLDD530 pKa = 4.23GSAYY534 pKa = 10.39GGTDD538 pKa = 3.18SGLDD542 pKa = 3.47PYY544 pKa = 11.58DD545 pKa = 4.83YY546 pKa = 10.76IALDD550 pKa = 3.98NDD552 pKa = 3.7TDD554 pKa = 4.16DD555 pKa = 5.96LGFSSDD561 pKa = 3.26NGATFTYY568 pKa = 10.27HH569 pKa = 6.23PTPDD573 pKa = 4.87GDD575 pKa = 4.35DD576 pKa = 3.54CDD578 pKa = 4.95EE579 pKa = 4.34NVTDD583 pKa = 4.86ILVNPKK589 pKa = 10.74GPMDD593 pKa = 4.36GAASGNTPSFQVRR606 pKa = 11.84FRR608 pKa = 11.84VQVEE612 pKa = 3.79
Molecular weight: 63.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A8ZS42|A8ZS42_DESOH Putative transcriptional regulator OS=Desulfococcus oleovorans (strain DSM 6200 / JCM 39069 / Hxd3) OX=96561 GN=Dole_0250 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.32QPSNVKK11 pKa = 10.02RR12 pKa = 11.84ARR14 pKa = 11.84KK15 pKa = 8.85HH16 pKa = 4.5GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.05QGRR28 pKa = 11.84SILKK32 pKa = 9.3RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.67GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.32QPSNVKK11 pKa = 10.02RR12 pKa = 11.84ARR14 pKa = 11.84KK15 pKa = 8.85HH16 pKa = 4.5GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.05QGRR28 pKa = 11.84SILKK32 pKa = 9.3RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.67GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1152799 |
34 |
12741 |
354.2 |
39.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.808 ± 0.053 | 1.33 ± 0.023 |
5.926 ± 0.057 | 5.963 ± 0.046 |
4.337 ± 0.03 | 7.829 ± 0.069 |
2.08 ± 0.02 | 6.015 ± 0.031 |
4.988 ± 0.057 | 9.493 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.757 ± 0.03 | 3.414 ± 0.034 |
4.673 ± 0.039 | 3.056 ± 0.023 |
5.909 ± 0.058 | 5.281 ± 0.031 |
5.64 ± 0.068 | 7.382 ± 0.039 |
1.155 ± 0.018 | 2.967 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
