
Lyngbya confervoides BDU141951
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Oscillatoriales; Oscillatoriaceae; Lyngbya; Lyngbya confervoides
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
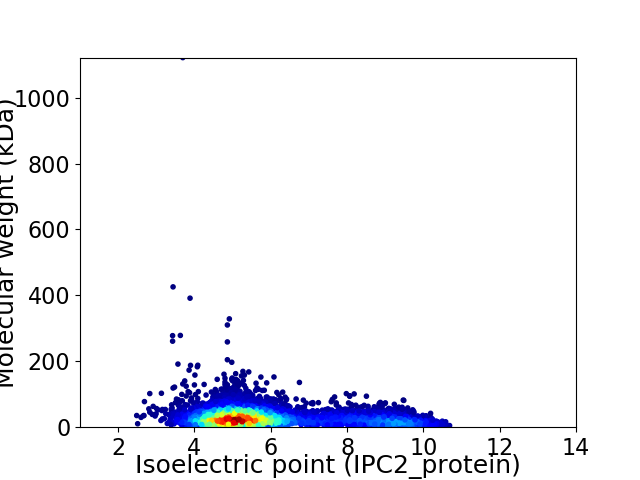
Virtual 2D-PAGE plot for 6060 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
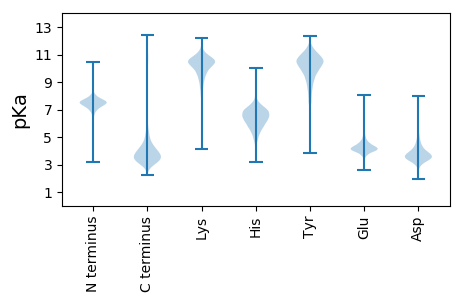
Protein with the lowest isoelectric point:
>tr|A0A0C1UPF6|A0A0C1UPF6_9CYAN DNA gyrase subunit B OS=Lyngbya confervoides BDU141951 OX=1574623 GN=gyrB PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 10.53LFVGLLSAGLVLAGGLSFASPSYY25 pKa = 9.33GQAVVATPRR34 pKa = 11.84SPALDD39 pKa = 3.28IQSTQIQYY47 pKa = 11.02LEE49 pKa = 4.14EE50 pKa = 5.65LDD52 pKa = 4.52LLVFEE57 pKa = 4.46QQLAGVVGATLPQSAGQLDD76 pKa = 4.16GAPVLGYY83 pKa = 9.39VFPTTLSPAAVGFGGVDD100 pKa = 4.43GILALAVTSHH110 pKa = 7.56PDD112 pKa = 3.75FDD114 pKa = 5.28DD115 pKa = 3.96TPLWDD120 pKa = 4.15EE121 pKa = 5.84DD122 pKa = 3.74NDD124 pKa = 3.86ADD126 pKa = 4.1YY127 pKa = 11.77ANDD130 pKa = 3.53GVIWHH135 pKa = 5.8THH137 pKa = 3.78WVVLTGDD144 pKa = 3.31NRR146 pKa = 11.84VPGGLSVAEE155 pKa = 4.6FAADD159 pKa = 3.65DD160 pKa = 4.32ASVVMPPTNPGMTIYY175 pKa = 10.56LDD177 pKa = 3.61SPGFSVVTDD186 pKa = 3.48QEE188 pKa = 4.47TLRR191 pKa = 11.84VLVPAQRR198 pKa = 11.84VDD200 pKa = 3.07HH201 pKa = 6.0NTDD204 pKa = 3.51FNFDD208 pKa = 3.13AVTAYY213 pKa = 9.25MQVNTSDD220 pKa = 3.33ASRR223 pKa = 11.84PTLGVYY229 pKa = 8.76EE230 pKa = 4.49VYY232 pKa = 10.71SVGSGDD238 pKa = 3.62LSLPYY243 pKa = 10.07GVRR246 pKa = 11.84HH247 pKa = 6.69DD248 pKa = 4.37ADD250 pKa = 3.27
MM1 pKa = 7.44KK2 pKa = 10.53LFVGLLSAGLVLAGGLSFASPSYY25 pKa = 9.33GQAVVATPRR34 pKa = 11.84SPALDD39 pKa = 3.28IQSTQIQYY47 pKa = 11.02LEE49 pKa = 4.14EE50 pKa = 5.65LDD52 pKa = 4.52LLVFEE57 pKa = 4.46QQLAGVVGATLPQSAGQLDD76 pKa = 4.16GAPVLGYY83 pKa = 9.39VFPTTLSPAAVGFGGVDD100 pKa = 4.43GILALAVTSHH110 pKa = 7.56PDD112 pKa = 3.75FDD114 pKa = 5.28DD115 pKa = 3.96TPLWDD120 pKa = 4.15EE121 pKa = 5.84DD122 pKa = 3.74NDD124 pKa = 3.86ADD126 pKa = 4.1YY127 pKa = 11.77ANDD130 pKa = 3.53GVIWHH135 pKa = 5.8THH137 pKa = 3.78WVVLTGDD144 pKa = 3.31NRR146 pKa = 11.84VPGGLSVAEE155 pKa = 4.6FAADD159 pKa = 3.65DD160 pKa = 4.32ASVVMPPTNPGMTIYY175 pKa = 10.56LDD177 pKa = 3.61SPGFSVVTDD186 pKa = 3.48QEE188 pKa = 4.47TLRR191 pKa = 11.84VLVPAQRR198 pKa = 11.84VDD200 pKa = 3.07HH201 pKa = 6.0NTDD204 pKa = 3.51FNFDD208 pKa = 3.13AVTAYY213 pKa = 9.25MQVNTSDD220 pKa = 3.33ASRR223 pKa = 11.84PTLGVYY229 pKa = 8.76EE230 pKa = 4.49VYY232 pKa = 10.71SVGSGDD238 pKa = 3.62LSLPYY243 pKa = 10.07GVRR246 pKa = 11.84HH247 pKa = 6.69DD248 pKa = 4.37ADD250 pKa = 3.27
Molecular weight: 26.35 kDa
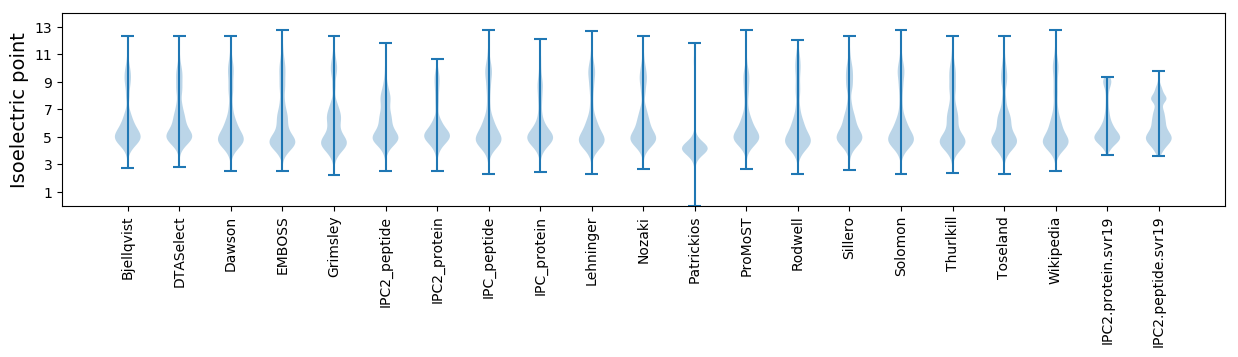
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C1USQ4|A0A0C1USQ4_9CYAN Multicopper oxidase OS=Lyngbya confervoides BDU141951 OX=1574623 GN=QQ91_30110 PE=4 SV=1
MM1 pKa = 7.38TEE3 pKa = 3.43ITRR6 pKa = 11.84IQAALRR12 pKa = 11.84PHH14 pKa = 6.92LSWHH18 pKa = 5.47GARR21 pKa = 11.84LTFLALFLVALFRR34 pKa = 11.84VEE36 pKa = 4.29TVNLEE41 pKa = 3.84RR42 pKa = 11.84LASVFANRR50 pKa = 11.84AQSASNHH57 pKa = 4.98KK58 pKa = 10.17RR59 pKa = 11.84LTRR62 pKa = 11.84FFRR65 pKa = 11.84KK66 pKa = 9.45FVIDD70 pKa = 3.87FEE72 pKa = 5.11EE73 pKa = 4.21IAQAVVSWSQIPEE86 pKa = 3.93PWTLSLDD93 pKa = 3.51RR94 pKa = 11.84TNWSFGTVNFNILMLGIVHH113 pKa = 6.89EE114 pKa = 5.01GVAFPVMWTMLDD126 pKa = 3.09KK127 pKa = 11.06RR128 pKa = 11.84GNSNSDD134 pKa = 2.72EE135 pKa = 4.45RR136 pKa = 11.84MDD138 pKa = 3.79LLEE141 pKa = 4.09RR142 pKa = 11.84FEE144 pKa = 5.07RR145 pKa = 11.84VFPNAQIRR153 pKa = 11.84CLTGDD158 pKa = 3.43RR159 pKa = 11.84EE160 pKa = 4.39FVGRR164 pKa = 11.84EE165 pKa = 3.49WCSFLLVPDD174 pKa = 5.78AIPFRR179 pKa = 11.84LRR181 pKa = 11.84LRR183 pKa = 11.84HH184 pKa = 5.91SDD186 pKa = 3.76RR187 pKa = 11.84ISSRR191 pKa = 11.84SGKK194 pKa = 8.75QRR196 pKa = 11.84QTGEE200 pKa = 3.96RR201 pKa = 11.84VFANLKK207 pKa = 10.06VGEE210 pKa = 3.98QRR212 pKa = 11.84HH213 pKa = 5.39LSGKK217 pKa = 7.9RR218 pKa = 11.84WVWGRR223 pKa = 11.84RR224 pKa = 11.84VYY226 pKa = 10.38IVATRR231 pKa = 11.84LEE233 pKa = 4.54DD234 pKa = 4.14GEE236 pKa = 4.72LLILATGHH244 pKa = 5.67QPQTAFADD252 pKa = 3.6YY253 pKa = 10.14RR254 pKa = 11.84LRR256 pKa = 11.84WGIEE260 pKa = 3.82TLFATLKK267 pKa = 9.6TRR269 pKa = 11.84GFNLEE274 pKa = 4.08STHH277 pKa = 5.89FCHH280 pKa = 7.72AEE282 pKa = 3.88RR283 pKa = 11.84LSKK286 pKa = 10.85LIALLALAFCWAMLTGLWQHH306 pKa = 5.51QQHH309 pKa = 7.42PIPLKK314 pKa = 9.02AHH316 pKa = 5.36GRR318 pKa = 11.84RR319 pKa = 11.84AKK321 pKa = 10.36SLFRR325 pKa = 11.84YY326 pKa = 9.9GCDD329 pKa = 3.22FLRR332 pKa = 11.84RR333 pKa = 11.84TFSDD337 pKa = 4.52FSLRR341 pKa = 11.84RR342 pKa = 11.84SEE344 pKa = 4.73FNQALKK350 pKa = 10.85LLSLYY355 pKa = 10.79
MM1 pKa = 7.38TEE3 pKa = 3.43ITRR6 pKa = 11.84IQAALRR12 pKa = 11.84PHH14 pKa = 6.92LSWHH18 pKa = 5.47GARR21 pKa = 11.84LTFLALFLVALFRR34 pKa = 11.84VEE36 pKa = 4.29TVNLEE41 pKa = 3.84RR42 pKa = 11.84LASVFANRR50 pKa = 11.84AQSASNHH57 pKa = 4.98KK58 pKa = 10.17RR59 pKa = 11.84LTRR62 pKa = 11.84FFRR65 pKa = 11.84KK66 pKa = 9.45FVIDD70 pKa = 3.87FEE72 pKa = 5.11EE73 pKa = 4.21IAQAVVSWSQIPEE86 pKa = 3.93PWTLSLDD93 pKa = 3.51RR94 pKa = 11.84TNWSFGTVNFNILMLGIVHH113 pKa = 6.89EE114 pKa = 5.01GVAFPVMWTMLDD126 pKa = 3.09KK127 pKa = 11.06RR128 pKa = 11.84GNSNSDD134 pKa = 2.72EE135 pKa = 4.45RR136 pKa = 11.84MDD138 pKa = 3.79LLEE141 pKa = 4.09RR142 pKa = 11.84FEE144 pKa = 5.07RR145 pKa = 11.84VFPNAQIRR153 pKa = 11.84CLTGDD158 pKa = 3.43RR159 pKa = 11.84EE160 pKa = 4.39FVGRR164 pKa = 11.84EE165 pKa = 3.49WCSFLLVPDD174 pKa = 5.78AIPFRR179 pKa = 11.84LRR181 pKa = 11.84LRR183 pKa = 11.84HH184 pKa = 5.91SDD186 pKa = 3.76RR187 pKa = 11.84ISSRR191 pKa = 11.84SGKK194 pKa = 8.75QRR196 pKa = 11.84QTGEE200 pKa = 3.96RR201 pKa = 11.84VFANLKK207 pKa = 10.06VGEE210 pKa = 3.98QRR212 pKa = 11.84HH213 pKa = 5.39LSGKK217 pKa = 7.9RR218 pKa = 11.84WVWGRR223 pKa = 11.84RR224 pKa = 11.84VYY226 pKa = 10.38IVATRR231 pKa = 11.84LEE233 pKa = 4.54DD234 pKa = 4.14GEE236 pKa = 4.72LLILATGHH244 pKa = 5.67QPQTAFADD252 pKa = 3.6YY253 pKa = 10.14RR254 pKa = 11.84LRR256 pKa = 11.84WGIEE260 pKa = 3.82TLFATLKK267 pKa = 9.6TRR269 pKa = 11.84GFNLEE274 pKa = 4.08STHH277 pKa = 5.89FCHH280 pKa = 7.72AEE282 pKa = 3.88RR283 pKa = 11.84LSKK286 pKa = 10.85LIALLALAFCWAMLTGLWQHH306 pKa = 5.51QQHH309 pKa = 7.42PIPLKK314 pKa = 9.02AHH316 pKa = 5.36GRR318 pKa = 11.84RR319 pKa = 11.84AKK321 pKa = 10.36SLFRR325 pKa = 11.84YY326 pKa = 9.9GCDD329 pKa = 3.22FLRR332 pKa = 11.84RR333 pKa = 11.84TFSDD337 pKa = 4.52FSLRR341 pKa = 11.84RR342 pKa = 11.84SEE344 pKa = 4.73FNQALKK350 pKa = 10.85LLSLYY355 pKa = 10.79
Molecular weight: 41.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1735983 |
28 |
10778 |
286.5 |
31.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.311 ± 0.041 | 0.945 ± 0.012 |
5.768 ± 0.04 | 6.014 ± 0.036 |
3.869 ± 0.023 | 7.49 ± 0.046 |
2.021 ± 0.02 | 5.563 ± 0.028 |
3.134 ± 0.032 | 10.717 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.211 ± 0.018 | 3.199 ± 0.031 |
5.179 ± 0.035 | 4.912 ± 0.03 |
5.806 ± 0.037 | 5.611 ± 0.027 |
5.886 ± 0.038 | 7.086 ± 0.024 |
1.502 ± 0.014 | 2.772 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
