
Macaca mulatta papillomavirus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; unclassified Papillomaviridae; primate papillomaviruses
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
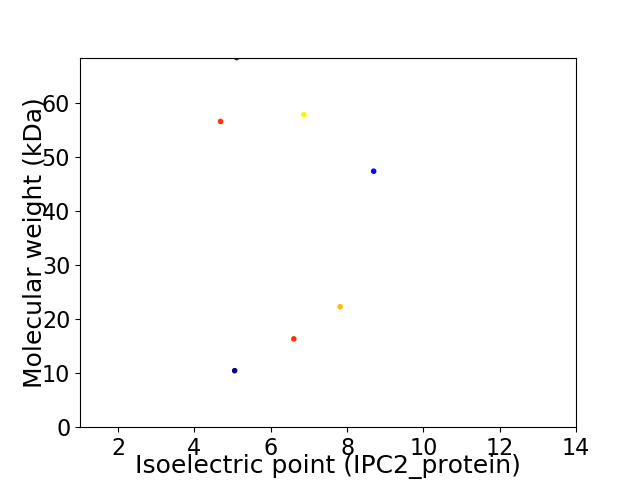
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A385AH05|A0A385AH05_9PAPI Replication protein E1 OS=Macaca mulatta papillomavirus 4 OX=2294152 GN=E1 PE=3 SV=1
MM1 pKa = 7.25HH2 pKa = 7.14RR3 pKa = 11.84AKK5 pKa = 10.25RR6 pKa = 11.84VKK8 pKa = 9.98RR9 pKa = 11.84DD10 pKa = 3.35SVQNIYY16 pKa = 9.4NQCKK20 pKa = 9.98ISGNCPEE27 pKa = 4.69DD28 pKa = 3.32VVNKK32 pKa = 10.0VEE34 pKa = 4.09QTTLADD40 pKa = 3.93RR41 pKa = 11.84LLKK44 pKa = 10.35ILGSIVYY51 pKa = 10.01FGGLGIGTGRR61 pKa = 11.84GTGGSTGYY69 pKa = 8.73TPLGAGSGRR78 pKa = 11.84VGAGGTVIRR87 pKa = 11.84PTVPVDD93 pKa = 3.65PIGPVDD99 pKa = 3.97ILPIDD104 pKa = 4.01SVDD107 pKa = 3.56PAASSIVPLSEE118 pKa = 4.63GGLPDD123 pKa = 3.73GAVVEE128 pKa = 4.86LEE130 pKa = 4.27PGGPDD135 pKa = 3.8LGAGDD140 pKa = 4.41IEE142 pKa = 4.85ILTEE146 pKa = 3.89TDD148 pKa = 4.08PISDD152 pKa = 3.4VTGRR156 pKa = 11.84GGHH159 pKa = 6.28PSISTTDD166 pKa = 3.02TDD168 pKa = 3.75VAVIDD173 pKa = 4.14VTPAPAGPRR182 pKa = 11.84RR183 pKa = 11.84TPTFSPIEE191 pKa = 4.14PSHH194 pKa = 7.4VRR196 pKa = 11.84IQASLSTSAEE206 pKa = 3.91PPNINIFIDD215 pKa = 3.73SSLSGQAVSVGEE227 pKa = 4.61EE228 pKa = 4.26IPLQDD233 pKa = 3.67IPLFDD238 pKa = 3.84EE239 pKa = 5.51LEE241 pKa = 4.49IEE243 pKa = 4.43EE244 pKa = 4.75PGPRR248 pKa = 11.84TSTPRR253 pKa = 11.84QVIEE257 pKa = 3.85RR258 pKa = 11.84AYY260 pKa = 8.59TRR262 pKa = 11.84ARR264 pKa = 11.84EE265 pKa = 4.06LYY267 pKa = 8.5NRR269 pKa = 11.84RR270 pKa = 11.84VAQVQTRR277 pKa = 11.84NVDD280 pKa = 3.94FLSRR284 pKa = 11.84PSRR287 pKa = 11.84LVQFEE292 pKa = 4.25FEE294 pKa = 4.2NPAFAEE300 pKa = 4.15EE301 pKa = 4.21DD302 pKa = 3.42VSLAFQQDD310 pKa = 3.65LQQIAAAPDD319 pKa = 3.67ADD321 pKa = 3.95FQDD324 pKa = 4.41VIRR327 pKa = 11.84LGRR330 pKa = 11.84PQLSEE335 pKa = 3.81TGEE338 pKa = 3.75GRR340 pKa = 11.84IRR342 pKa = 11.84YY343 pKa = 8.83SRR345 pKa = 11.84AGRR348 pKa = 11.84RR349 pKa = 11.84GTIRR353 pKa = 11.84TRR355 pKa = 11.84SGVQIGQHH363 pKa = 3.59VHH365 pKa = 6.48FYY367 pKa = 10.88YY368 pKa = 10.83DD369 pKa = 3.76FSTIEE374 pKa = 3.84NADD377 pKa = 4.06AIEE380 pKa = 4.1LQPLGEE386 pKa = 4.26VTGDD390 pKa = 3.35LTLIDD395 pKa = 4.26AQAEE399 pKa = 4.47SSFVDD404 pKa = 3.87SVAVDD409 pKa = 3.44SASEE413 pKa = 4.26HH414 pKa = 6.51IYY416 pKa = 10.84SDD418 pKa = 3.6EE419 pKa = 4.2ALLDD423 pKa = 4.13PYY425 pKa = 11.3VEE427 pKa = 4.61SFDD430 pKa = 4.04NSHH433 pKa = 6.67LVLSVGRR440 pKa = 11.84RR441 pKa = 11.84TDD443 pKa = 2.97SFTVPSIPPGISLKK457 pKa = 10.83VFVDD461 pKa = 4.29DD462 pKa = 4.59YY463 pKa = 11.62GADD466 pKa = 4.17LIVSYY471 pKa = 9.48PQSQDD476 pKa = 2.58HH477 pKa = 7.01TIPHH481 pKa = 6.95PLFPFGPVEE490 pKa = 3.97PVVVFPTSDD499 pKa = 3.05GSDD502 pKa = 3.71YY503 pKa = 11.15LLHH506 pKa = 6.99PSLLKK511 pKa = 9.89RR512 pKa = 11.84RR513 pKa = 11.84RR514 pKa = 11.84RR515 pKa = 11.84KK516 pKa = 9.58RR517 pKa = 11.84RR518 pKa = 11.84YY519 pKa = 9.65SDD521 pKa = 3.18TFF523 pKa = 3.61
MM1 pKa = 7.25HH2 pKa = 7.14RR3 pKa = 11.84AKK5 pKa = 10.25RR6 pKa = 11.84VKK8 pKa = 9.98RR9 pKa = 11.84DD10 pKa = 3.35SVQNIYY16 pKa = 9.4NQCKK20 pKa = 9.98ISGNCPEE27 pKa = 4.69DD28 pKa = 3.32VVNKK32 pKa = 10.0VEE34 pKa = 4.09QTTLADD40 pKa = 3.93RR41 pKa = 11.84LLKK44 pKa = 10.35ILGSIVYY51 pKa = 10.01FGGLGIGTGRR61 pKa = 11.84GTGGSTGYY69 pKa = 8.73TPLGAGSGRR78 pKa = 11.84VGAGGTVIRR87 pKa = 11.84PTVPVDD93 pKa = 3.65PIGPVDD99 pKa = 3.97ILPIDD104 pKa = 4.01SVDD107 pKa = 3.56PAASSIVPLSEE118 pKa = 4.63GGLPDD123 pKa = 3.73GAVVEE128 pKa = 4.86LEE130 pKa = 4.27PGGPDD135 pKa = 3.8LGAGDD140 pKa = 4.41IEE142 pKa = 4.85ILTEE146 pKa = 3.89TDD148 pKa = 4.08PISDD152 pKa = 3.4VTGRR156 pKa = 11.84GGHH159 pKa = 6.28PSISTTDD166 pKa = 3.02TDD168 pKa = 3.75VAVIDD173 pKa = 4.14VTPAPAGPRR182 pKa = 11.84RR183 pKa = 11.84TPTFSPIEE191 pKa = 4.14PSHH194 pKa = 7.4VRR196 pKa = 11.84IQASLSTSAEE206 pKa = 3.91PPNINIFIDD215 pKa = 3.73SSLSGQAVSVGEE227 pKa = 4.61EE228 pKa = 4.26IPLQDD233 pKa = 3.67IPLFDD238 pKa = 3.84EE239 pKa = 5.51LEE241 pKa = 4.49IEE243 pKa = 4.43EE244 pKa = 4.75PGPRR248 pKa = 11.84TSTPRR253 pKa = 11.84QVIEE257 pKa = 3.85RR258 pKa = 11.84AYY260 pKa = 8.59TRR262 pKa = 11.84ARR264 pKa = 11.84EE265 pKa = 4.06LYY267 pKa = 8.5NRR269 pKa = 11.84RR270 pKa = 11.84VAQVQTRR277 pKa = 11.84NVDD280 pKa = 3.94FLSRR284 pKa = 11.84PSRR287 pKa = 11.84LVQFEE292 pKa = 4.25FEE294 pKa = 4.2NPAFAEE300 pKa = 4.15EE301 pKa = 4.21DD302 pKa = 3.42VSLAFQQDD310 pKa = 3.65LQQIAAAPDD319 pKa = 3.67ADD321 pKa = 3.95FQDD324 pKa = 4.41VIRR327 pKa = 11.84LGRR330 pKa = 11.84PQLSEE335 pKa = 3.81TGEE338 pKa = 3.75GRR340 pKa = 11.84IRR342 pKa = 11.84YY343 pKa = 8.83SRR345 pKa = 11.84AGRR348 pKa = 11.84RR349 pKa = 11.84GTIRR353 pKa = 11.84TRR355 pKa = 11.84SGVQIGQHH363 pKa = 3.59VHH365 pKa = 6.48FYY367 pKa = 10.88YY368 pKa = 10.83DD369 pKa = 3.76FSTIEE374 pKa = 3.84NADD377 pKa = 4.06AIEE380 pKa = 4.1LQPLGEE386 pKa = 4.26VTGDD390 pKa = 3.35LTLIDD395 pKa = 4.26AQAEE399 pKa = 4.47SSFVDD404 pKa = 3.87SVAVDD409 pKa = 3.44SASEE413 pKa = 4.26HH414 pKa = 6.51IYY416 pKa = 10.84SDD418 pKa = 3.6EE419 pKa = 4.2ALLDD423 pKa = 4.13PYY425 pKa = 11.3VEE427 pKa = 4.61SFDD430 pKa = 4.04NSHH433 pKa = 6.67LVLSVGRR440 pKa = 11.84RR441 pKa = 11.84TDD443 pKa = 2.97SFTVPSIPPGISLKK457 pKa = 10.83VFVDD461 pKa = 4.29DD462 pKa = 4.59YY463 pKa = 11.62GADD466 pKa = 4.17LIVSYY471 pKa = 9.48PQSQDD476 pKa = 2.58HH477 pKa = 7.01TIPHH481 pKa = 6.95PLFPFGPVEE490 pKa = 3.97PVVVFPTSDD499 pKa = 3.05GSDD502 pKa = 3.71YY503 pKa = 11.15LLHH506 pKa = 6.99PSLLKK511 pKa = 9.89RR512 pKa = 11.84RR513 pKa = 11.84RR514 pKa = 11.84RR515 pKa = 11.84KK516 pKa = 9.58RR517 pKa = 11.84RR518 pKa = 11.84YY519 pKa = 9.65SDD521 pKa = 3.18TFF523 pKa = 3.61
Molecular weight: 56.54 kDa
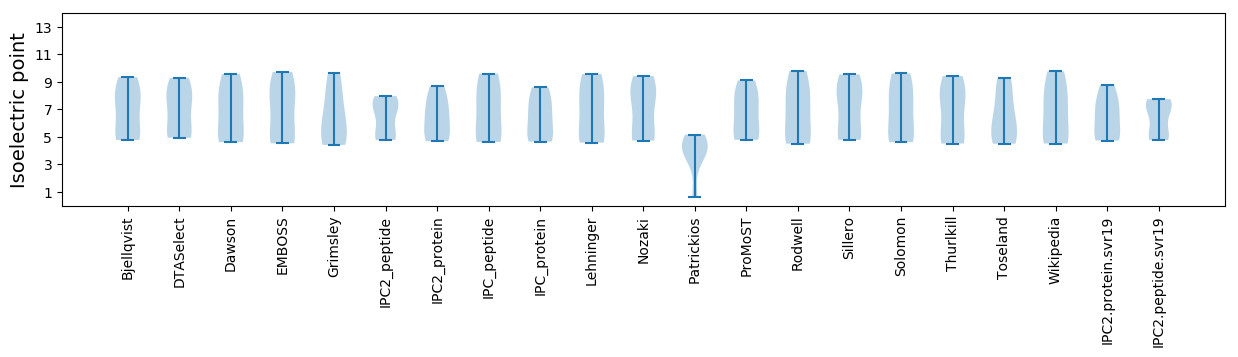
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A385AHG8|A0A385AHG8_9PAPI E4 OS=Macaca mulatta papillomavirus 4 OX=2294152 GN=E4 PE=4 SV=1
MM1 pKa = 7.03EE2 pKa = 4.64TLTEE6 pKa = 3.97RR7 pKa = 11.84FEE9 pKa = 4.59TVQEE13 pKa = 4.17ALLKK17 pKa = 10.45LYY19 pKa = 10.27EE20 pKa = 4.47SEE22 pKa = 4.22ATDD25 pKa = 2.95IDD27 pKa = 4.01TQIEE31 pKa = 4.2HH32 pKa = 6.76WNLIRR37 pKa = 11.84KK38 pKa = 8.46EE39 pKa = 4.11NILLHH44 pKa = 5.09YY45 pKa = 9.79ARR47 pKa = 11.84KK48 pKa = 10.2NGVSKK53 pKa = 11.06LGLHH57 pKa = 6.05TVPQLTVSEE66 pKa = 4.85HH67 pKa = 5.53NAKK70 pKa = 9.85QAIKK74 pKa = 9.94MVLLLQKK81 pKa = 10.75LKK83 pKa = 10.77DD84 pKa = 3.57SEE86 pKa = 4.44YY87 pKa = 11.16GKK89 pKa = 10.29EE90 pKa = 3.7RR91 pKa = 11.84WTLTDD96 pKa = 3.08TSLEE100 pKa = 4.15LFNTPPKK107 pKa = 10.67NCLKK111 pKa = 10.76KK112 pKa = 10.87GGFTVDD118 pKa = 3.05VLYY121 pKa = 11.05DD122 pKa = 3.52NEE124 pKa = 4.02EE125 pKa = 4.37SKK127 pKa = 10.84RR128 pKa = 11.84FPYY131 pKa = 9.05TAWTYY136 pKa = 10.84IYY138 pKa = 10.32YY139 pKa = 10.86LNDD142 pKa = 2.91NDD144 pKa = 3.62EE145 pKa = 4.27WEE147 pKa = 4.47KK148 pKa = 11.22VPGKK152 pKa = 10.4ADD154 pKa = 3.47YY155 pKa = 10.82YY156 pKa = 9.77GLYY159 pKa = 10.15YY160 pKa = 10.44EE161 pKa = 4.57EE162 pKa = 5.2SGHH165 pKa = 5.86KK166 pKa = 9.72RR167 pKa = 11.84YY168 pKa = 9.85YY169 pKa = 11.05SKK171 pKa = 11.12FEE173 pKa = 3.96EE174 pKa = 4.93DD175 pKa = 3.0AAQYY179 pKa = 9.6GQTGIWEE186 pKa = 3.96VRR188 pKa = 11.84YY189 pKa = 10.26KK190 pKa = 11.27NNIISAPVTSSTKK203 pKa = 10.58APGSGSAAGVPARR216 pKa = 11.84PTGHH220 pKa = 6.84TEE222 pKa = 3.21TDD224 pKa = 2.95AQTRR228 pKa = 11.84GGRR231 pKa = 11.84RR232 pKa = 11.84KK233 pKa = 9.15VHH235 pKa = 5.99FAPRR239 pKa = 11.84GCTPSTNLSSPLRR252 pKa = 11.84EE253 pKa = 3.73EE254 pKa = 4.13AKK256 pKa = 10.02RR257 pKa = 11.84PRR259 pKa = 11.84GGRR262 pKa = 11.84VGRR265 pKa = 11.84GGQQRR270 pKa = 11.84GQGEE274 pKa = 4.17QGPPQRR280 pKa = 11.84GEE282 pKa = 4.08SPSSKK287 pKa = 9.56RR288 pKa = 11.84RR289 pKa = 11.84RR290 pKa = 11.84IDD292 pKa = 3.12IPDD295 pKa = 3.63SSTGRR300 pKa = 11.84PTFPSPSEE308 pKa = 3.7VGSRR312 pKa = 11.84HH313 pKa = 5.63RR314 pKa = 11.84SVEE317 pKa = 3.69GGHH320 pKa = 6.98RR321 pKa = 11.84SRR323 pKa = 11.84LQQLQDD329 pKa = 3.45EE330 pKa = 4.77ARR332 pKa = 11.84DD333 pKa = 3.76PPVVIIKK340 pKa = 8.65GIPNILKK347 pKa = 9.75CWRR350 pKa = 11.84NRR352 pKa = 11.84TKK354 pKa = 10.65QKK356 pKa = 10.4NSRR359 pKa = 11.84LFTCISSVFTWKK371 pKa = 10.62DD372 pKa = 3.2DD373 pKa = 4.43DD374 pKa = 5.97DD375 pKa = 4.38NACTGSGAKK384 pKa = 10.16GRR386 pKa = 11.84MLVAFDD392 pKa = 3.4TTRR395 pKa = 11.84QRR397 pKa = 11.84DD398 pKa = 3.41IFLKK402 pKa = 9.88TVHH405 pKa = 6.24LPKK408 pKa = 10.09GTSYY412 pKa = 11.27DD413 pKa = 3.39IGSLGSLL420 pKa = 3.72
MM1 pKa = 7.03EE2 pKa = 4.64TLTEE6 pKa = 3.97RR7 pKa = 11.84FEE9 pKa = 4.59TVQEE13 pKa = 4.17ALLKK17 pKa = 10.45LYY19 pKa = 10.27EE20 pKa = 4.47SEE22 pKa = 4.22ATDD25 pKa = 2.95IDD27 pKa = 4.01TQIEE31 pKa = 4.2HH32 pKa = 6.76WNLIRR37 pKa = 11.84KK38 pKa = 8.46EE39 pKa = 4.11NILLHH44 pKa = 5.09YY45 pKa = 9.79ARR47 pKa = 11.84KK48 pKa = 10.2NGVSKK53 pKa = 11.06LGLHH57 pKa = 6.05TVPQLTVSEE66 pKa = 4.85HH67 pKa = 5.53NAKK70 pKa = 9.85QAIKK74 pKa = 9.94MVLLLQKK81 pKa = 10.75LKK83 pKa = 10.77DD84 pKa = 3.57SEE86 pKa = 4.44YY87 pKa = 11.16GKK89 pKa = 10.29EE90 pKa = 3.7RR91 pKa = 11.84WTLTDD96 pKa = 3.08TSLEE100 pKa = 4.15LFNTPPKK107 pKa = 10.67NCLKK111 pKa = 10.76KK112 pKa = 10.87GGFTVDD118 pKa = 3.05VLYY121 pKa = 11.05DD122 pKa = 3.52NEE124 pKa = 4.02EE125 pKa = 4.37SKK127 pKa = 10.84RR128 pKa = 11.84FPYY131 pKa = 9.05TAWTYY136 pKa = 10.84IYY138 pKa = 10.32YY139 pKa = 10.86LNDD142 pKa = 2.91NDD144 pKa = 3.62EE145 pKa = 4.27WEE147 pKa = 4.47KK148 pKa = 11.22VPGKK152 pKa = 10.4ADD154 pKa = 3.47YY155 pKa = 10.82YY156 pKa = 9.77GLYY159 pKa = 10.15YY160 pKa = 10.44EE161 pKa = 4.57EE162 pKa = 5.2SGHH165 pKa = 5.86KK166 pKa = 9.72RR167 pKa = 11.84YY168 pKa = 9.85YY169 pKa = 11.05SKK171 pKa = 11.12FEE173 pKa = 3.96EE174 pKa = 4.93DD175 pKa = 3.0AAQYY179 pKa = 9.6GQTGIWEE186 pKa = 3.96VRR188 pKa = 11.84YY189 pKa = 10.26KK190 pKa = 11.27NNIISAPVTSSTKK203 pKa = 10.58APGSGSAAGVPARR216 pKa = 11.84PTGHH220 pKa = 6.84TEE222 pKa = 3.21TDD224 pKa = 2.95AQTRR228 pKa = 11.84GGRR231 pKa = 11.84RR232 pKa = 11.84KK233 pKa = 9.15VHH235 pKa = 5.99FAPRR239 pKa = 11.84GCTPSTNLSSPLRR252 pKa = 11.84EE253 pKa = 3.73EE254 pKa = 4.13AKK256 pKa = 10.02RR257 pKa = 11.84PRR259 pKa = 11.84GGRR262 pKa = 11.84VGRR265 pKa = 11.84GGQQRR270 pKa = 11.84GQGEE274 pKa = 4.17QGPPQRR280 pKa = 11.84GEE282 pKa = 4.08SPSSKK287 pKa = 9.56RR288 pKa = 11.84RR289 pKa = 11.84RR290 pKa = 11.84IDD292 pKa = 3.12IPDD295 pKa = 3.63SSTGRR300 pKa = 11.84PTFPSPSEE308 pKa = 3.7VGSRR312 pKa = 11.84HH313 pKa = 5.63RR314 pKa = 11.84SVEE317 pKa = 3.69GGHH320 pKa = 6.98RR321 pKa = 11.84SRR323 pKa = 11.84LQQLQDD329 pKa = 3.45EE330 pKa = 4.77ARR332 pKa = 11.84DD333 pKa = 3.76PPVVIIKK340 pKa = 8.65GIPNILKK347 pKa = 9.75CWRR350 pKa = 11.84NRR352 pKa = 11.84TKK354 pKa = 10.65QKK356 pKa = 10.4NSRR359 pKa = 11.84LFTCISSVFTWKK371 pKa = 10.62DD372 pKa = 3.2DD373 pKa = 4.43DD374 pKa = 5.97DD375 pKa = 4.38NACTGSGAKK384 pKa = 10.16GRR386 pKa = 11.84MLVAFDD392 pKa = 3.4TTRR395 pKa = 11.84QRR397 pKa = 11.84DD398 pKa = 3.41IFLKK402 pKa = 9.88TVHH405 pKa = 6.24LPKK408 pKa = 10.09GTSYY412 pKa = 11.27DD413 pKa = 3.39IGSLGSLL420 pKa = 3.72
Molecular weight: 47.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2482 |
94 |
598 |
354.6 |
39.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.963 ± 0.313 | 2.176 ± 0.722 |
6.326 ± 0.409 | 6.527 ± 0.614 |
4.19 ± 0.525 | 6.487 ± 0.867 |
1.934 ± 0.248 | 4.593 ± 0.583 |
5.479 ± 1.024 | 9.065 ± 0.931 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.652 ± 0.562 | 4.069 ± 0.544 |
6.608 ± 0.702 | 4.432 ± 0.14 |
6.205 ± 0.721 | 7.816 ± 0.571 |
5.923 ± 0.506 | 5.761 ± 0.954 |
1.249 ± 0.312 | 3.546 ± 0.327 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
