
endosymbiont of unidentified scaly snail isolate Monju
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; unclassified Gammaproteobacteria
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
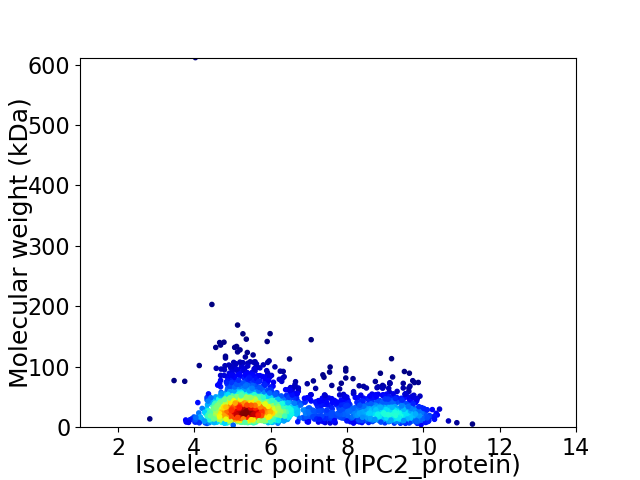
Virtual 2D-PAGE plot for 2228 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S6BY56|S6BY56_9GAMM Glutaredoxin OS=endosymbiont of unidentified scaly snail isolate Monju OX=1248727 GN=EBS_1184 PE=4 SV=1
MM1 pKa = 7.77LILDD5 pKa = 3.88FSGSVRR11 pKa = 11.84GQNLADD17 pKa = 3.51MQAAVISMLDD27 pKa = 3.16AYY29 pKa = 11.2NNAGYY34 pKa = 10.74AVVQLVTFSGNANIPADD51 pKa = 4.08GGWISVADD59 pKa = 4.04AKK61 pKa = 11.16AYY63 pKa = 10.1INGLTDD69 pKa = 3.76ADD71 pKa = 3.85MGGSTNYY78 pKa = 10.23DD79 pKa = 3.12AALAAAQSAFAEE91 pKa = 4.41AAGKK95 pKa = 10.22IPGAKK100 pKa = 9.77NIAYY104 pKa = 8.84FLSDD108 pKa = 3.88GSPTSGNGSIGIDD121 pKa = 3.6PAEE124 pKa = 3.79QAAWEE129 pKa = 4.1NFLTNNAIDD138 pKa = 3.53SHH140 pKa = 7.62AVGFGGASTTEE151 pKa = 4.29LEE153 pKa = 4.55PIAYY157 pKa = 9.98NGIDD161 pKa = 3.66GTEE164 pKa = 4.2CPALDD169 pKa = 3.87ATTAGANLTQVLLDD183 pKa = 3.87TVAQTVPGNLFGSLATGGFGADD205 pKa = 3.25GAGIVTSLTVGGVTYY220 pKa = 10.12AYY222 pKa = 10.46DD223 pKa = 3.86SNNDD227 pKa = 3.85TITGGSSTLNGHH239 pKa = 5.82QLTVTTSQGGILSVNMLTGEE259 pKa = 4.14YY260 pKa = 9.61TYY262 pKa = 11.19LADD265 pKa = 3.5PTFTISYY272 pKa = 9.65NEE274 pKa = 3.65IVAYY278 pKa = 10.07ALQDD282 pKa = 3.46ADD284 pKa = 4.07GDD286 pKa = 4.26ATSGTLTLNVARR298 pKa = 11.84DD299 pKa = 3.97VKK301 pKa = 10.49PVPTLLDD308 pKa = 3.33NTADD312 pKa = 3.57VYY314 pKa = 10.28EE315 pKa = 4.56AAMSTGTNPDD325 pKa = 2.89STAEE329 pKa = 4.1VATGNILSDD338 pKa = 3.57DD339 pKa = 4.48TIPAGLSLSNVSIAGGATVINGNTITVTTAEE370 pKa = 4.42GNTLVVDD377 pKa = 5.35KK378 pKa = 10.49ITGDD382 pKa = 3.41YY383 pKa = 10.33TYY385 pKa = 10.97TLNNPVKK392 pKa = 10.16HH393 pKa = 6.33LLFSATGNQVTLANDD408 pKa = 3.84TFTGGVLDD416 pKa = 4.6GWTGTNVSNKK426 pKa = 9.77NDD428 pKa = 3.24WLRR431 pKa = 11.84IDD433 pKa = 3.51GRR435 pKa = 11.84GDD437 pKa = 3.18VATKK441 pKa = 10.23TFDD444 pKa = 3.53FGQSYY449 pKa = 11.34ANQTVHH455 pKa = 5.08VTFDD459 pKa = 3.56FKK461 pKa = 11.77ANDD464 pKa = 3.03KK465 pKa = 10.06WDD467 pKa = 4.03ANTSDD472 pKa = 4.07SFRR475 pKa = 11.84MAVNGVEE482 pKa = 4.15ISNVPYY488 pKa = 10.59GKK490 pKa = 9.91NATDD494 pKa = 3.47TYY496 pKa = 11.19SFDD499 pKa = 3.49VTLDD503 pKa = 3.59ASGKK507 pKa = 10.63AYY509 pKa = 10.73FEE511 pKa = 4.54LTASTNSNKK520 pKa = 9.77EE521 pKa = 3.72DD522 pKa = 3.55AFVDD526 pKa = 3.43NFKK529 pKa = 9.36ITGPQLVPTPTDD541 pKa = 3.29VLVDD545 pKa = 3.64SFTYY549 pKa = 9.97TVTDD553 pKa = 4.21LGGTAYY559 pKa = 10.51NSKK562 pKa = 10.75LNVSIHH568 pKa = 7.3DD569 pKa = 4.32DD570 pKa = 3.6APIATTQNQQINVPQQDD587 pKa = 3.59TNLMVILDD595 pKa = 4.8LSGSMQGSRR604 pKa = 11.84LAAARR609 pKa = 11.84TAISNLIDD617 pKa = 3.42TYY619 pKa = 11.51NGYY622 pKa = 11.3GDD624 pKa = 3.8VAVKK628 pKa = 10.38LVTFSTLAQEE638 pKa = 4.37KK639 pKa = 8.44TSYY642 pKa = 10.15WMTASEE648 pKa = 4.52AKK650 pKa = 10.51AILATLSASGWTNYY664 pKa = 7.91DD665 pKa = 3.14TALAQAIQSWDD676 pKa = 3.4DD677 pKa = 3.4GSRR680 pKa = 11.84ITTPPSGGVIQNVAYY695 pKa = 9.65FISDD699 pKa = 3.8GQPNMNDD706 pKa = 3.02GDD708 pKa = 4.28TTVLANSNAGGTSGADD724 pKa = 3.13AGIQAAEE731 pKa = 4.14EE732 pKa = 4.47STT734 pKa = 3.51
MM1 pKa = 7.77LILDD5 pKa = 3.88FSGSVRR11 pKa = 11.84GQNLADD17 pKa = 3.51MQAAVISMLDD27 pKa = 3.16AYY29 pKa = 11.2NNAGYY34 pKa = 10.74AVVQLVTFSGNANIPADD51 pKa = 4.08GGWISVADD59 pKa = 4.04AKK61 pKa = 11.16AYY63 pKa = 10.1INGLTDD69 pKa = 3.76ADD71 pKa = 3.85MGGSTNYY78 pKa = 10.23DD79 pKa = 3.12AALAAAQSAFAEE91 pKa = 4.41AAGKK95 pKa = 10.22IPGAKK100 pKa = 9.77NIAYY104 pKa = 8.84FLSDD108 pKa = 3.88GSPTSGNGSIGIDD121 pKa = 3.6PAEE124 pKa = 3.79QAAWEE129 pKa = 4.1NFLTNNAIDD138 pKa = 3.53SHH140 pKa = 7.62AVGFGGASTTEE151 pKa = 4.29LEE153 pKa = 4.55PIAYY157 pKa = 9.98NGIDD161 pKa = 3.66GTEE164 pKa = 4.2CPALDD169 pKa = 3.87ATTAGANLTQVLLDD183 pKa = 3.87TVAQTVPGNLFGSLATGGFGADD205 pKa = 3.25GAGIVTSLTVGGVTYY220 pKa = 10.12AYY222 pKa = 10.46DD223 pKa = 3.86SNNDD227 pKa = 3.85TITGGSSTLNGHH239 pKa = 5.82QLTVTTSQGGILSVNMLTGEE259 pKa = 4.14YY260 pKa = 9.61TYY262 pKa = 11.19LADD265 pKa = 3.5PTFTISYY272 pKa = 9.65NEE274 pKa = 3.65IVAYY278 pKa = 10.07ALQDD282 pKa = 3.46ADD284 pKa = 4.07GDD286 pKa = 4.26ATSGTLTLNVARR298 pKa = 11.84DD299 pKa = 3.97VKK301 pKa = 10.49PVPTLLDD308 pKa = 3.33NTADD312 pKa = 3.57VYY314 pKa = 10.28EE315 pKa = 4.56AAMSTGTNPDD325 pKa = 2.89STAEE329 pKa = 4.1VATGNILSDD338 pKa = 3.57DD339 pKa = 4.48TIPAGLSLSNVSIAGGATVINGNTITVTTAEE370 pKa = 4.42GNTLVVDD377 pKa = 5.35KK378 pKa = 10.49ITGDD382 pKa = 3.41YY383 pKa = 10.33TYY385 pKa = 10.97TLNNPVKK392 pKa = 10.16HH393 pKa = 6.33LLFSATGNQVTLANDD408 pKa = 3.84TFTGGVLDD416 pKa = 4.6GWTGTNVSNKK426 pKa = 9.77NDD428 pKa = 3.24WLRR431 pKa = 11.84IDD433 pKa = 3.51GRR435 pKa = 11.84GDD437 pKa = 3.18VATKK441 pKa = 10.23TFDD444 pKa = 3.53FGQSYY449 pKa = 11.34ANQTVHH455 pKa = 5.08VTFDD459 pKa = 3.56FKK461 pKa = 11.77ANDD464 pKa = 3.03KK465 pKa = 10.06WDD467 pKa = 4.03ANTSDD472 pKa = 4.07SFRR475 pKa = 11.84MAVNGVEE482 pKa = 4.15ISNVPYY488 pKa = 10.59GKK490 pKa = 9.91NATDD494 pKa = 3.47TYY496 pKa = 11.19SFDD499 pKa = 3.49VTLDD503 pKa = 3.59ASGKK507 pKa = 10.63AYY509 pKa = 10.73FEE511 pKa = 4.54LTASTNSNKK520 pKa = 9.77EE521 pKa = 3.72DD522 pKa = 3.55AFVDD526 pKa = 3.43NFKK529 pKa = 9.36ITGPQLVPTPTDD541 pKa = 3.29VLVDD545 pKa = 3.64SFTYY549 pKa = 9.97TVTDD553 pKa = 4.21LGGTAYY559 pKa = 10.51NSKK562 pKa = 10.75LNVSIHH568 pKa = 7.3DD569 pKa = 4.32DD570 pKa = 3.6APIATTQNQQINVPQQDD587 pKa = 3.59TNLMVILDD595 pKa = 4.8LSGSMQGSRR604 pKa = 11.84LAAARR609 pKa = 11.84TAISNLIDD617 pKa = 3.42TYY619 pKa = 11.51NGYY622 pKa = 11.3GDD624 pKa = 3.8VAVKK628 pKa = 10.38LVTFSTLAQEE638 pKa = 4.37KK639 pKa = 8.44TSYY642 pKa = 10.15WMTASEE648 pKa = 4.52AKK650 pKa = 10.51AILATLSASGWTNYY664 pKa = 7.91DD665 pKa = 3.14TALAQAIQSWDD676 pKa = 3.4DD677 pKa = 3.4GSRR680 pKa = 11.84ITTPPSGGVIQNVAYY695 pKa = 9.65FISDD699 pKa = 3.8GQPNMNDD706 pKa = 3.02GDD708 pKa = 4.28TTVLANSNAGGTSGADD724 pKa = 3.13AGIQAAEE731 pKa = 4.14EE732 pKa = 4.47STT734 pKa = 3.51
Molecular weight: 75.97 kDa
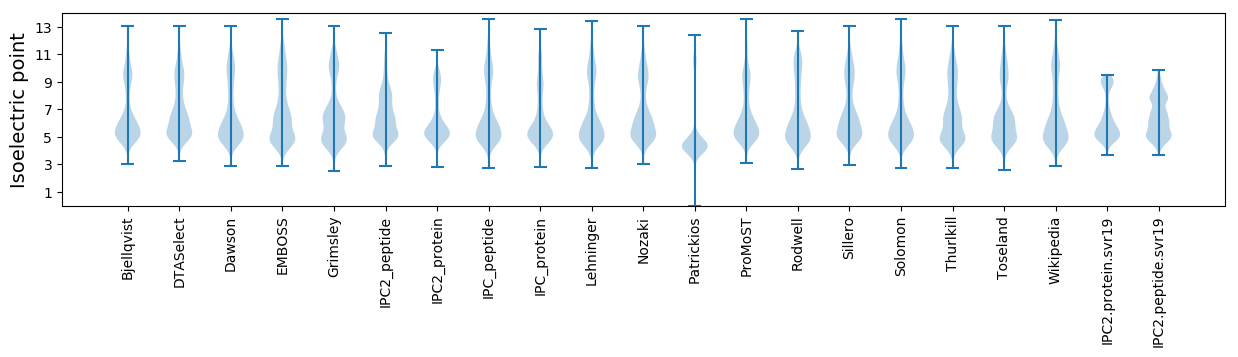
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S6BA54|S6BA54_9GAMM Phosphopantetheine adenylyltransferase OS=endosymbiont of unidentified scaly snail isolate Monju OX=1248727 GN=coaD PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84AGRR28 pKa = 11.84KK29 pKa = 8.84IINARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.53GRR39 pKa = 11.84ARR41 pKa = 11.84LAPP44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84AGRR28 pKa = 11.84KK29 pKa = 8.84IINARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.53GRR39 pKa = 11.84ARR41 pKa = 11.84LAPP44 pKa = 4.03
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
708729 |
30 |
5867 |
318.1 |
35.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.01 ± 0.069 | 0.992 ± 0.022 |
5.869 ± 0.056 | 6.919 ± 0.063 |
3.362 ± 0.032 | 8.144 ± 0.059 |
2.549 ± 0.028 | 4.862 ± 0.041 |
2.869 ± 0.049 | 11.407 ± 0.071 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.321 ± 0.024 | 2.417 ± 0.029 |
5.214 ± 0.037 | 3.729 ± 0.032 |
8.275 ± 0.059 | 4.505 ± 0.035 |
4.524 ± 0.038 | 7.293 ± 0.049 |
1.403 ± 0.026 | 2.336 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
