
Capybara microvirus Cap3_SP_416
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
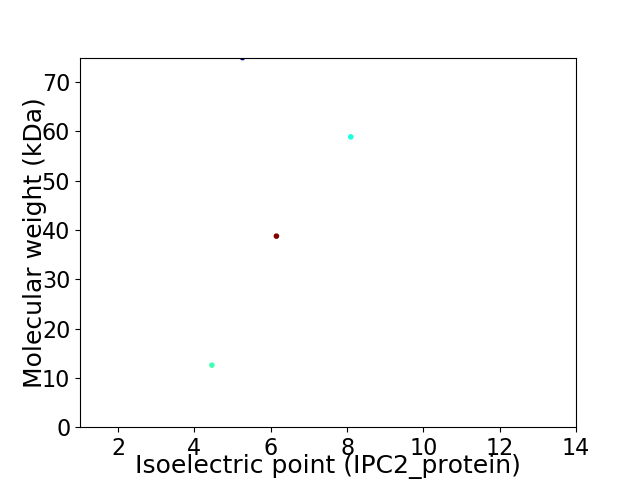
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W527|A0A4P8W527_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_416 OX=2585448 PE=4 SV=1
MM1 pKa = 7.13FRR3 pKa = 11.84KK4 pKa = 9.05IQNFIPEE11 pKa = 4.15PSSEE15 pKa = 3.61IFYY18 pKa = 11.22NEE20 pKa = 3.73DD21 pKa = 3.03EE22 pKa = 4.2EE23 pKa = 6.38RR24 pKa = 11.84YY25 pKa = 10.51VLVDD29 pKa = 3.74DD30 pKa = 4.21SPIRR34 pKa = 11.84VISSTPIDD42 pKa = 3.76ACNGKK47 pKa = 9.69LPASIPHH54 pKa = 5.52FQEE57 pKa = 4.08FTLTTLLNAGVKK69 pKa = 10.08LEE71 pKa = 3.89QLNTVLLRR79 pKa = 11.84PSVQQFEE86 pKa = 4.31QSFTDD91 pKa = 5.16FFTQTSNEE99 pKa = 4.05FVNHH103 pKa = 5.77YY104 pKa = 10.29EE105 pKa = 4.01SDD107 pKa = 3.91NII109 pKa = 4.19
MM1 pKa = 7.13FRR3 pKa = 11.84KK4 pKa = 9.05IQNFIPEE11 pKa = 4.15PSSEE15 pKa = 3.61IFYY18 pKa = 11.22NEE20 pKa = 3.73DD21 pKa = 3.03EE22 pKa = 4.2EE23 pKa = 6.38RR24 pKa = 11.84YY25 pKa = 10.51VLVDD29 pKa = 3.74DD30 pKa = 4.21SPIRR34 pKa = 11.84VISSTPIDD42 pKa = 3.76ACNGKK47 pKa = 9.69LPASIPHH54 pKa = 5.52FQEE57 pKa = 4.08FTLTTLLNAGVKK69 pKa = 10.08LEE71 pKa = 3.89QLNTVLLRR79 pKa = 11.84PSVQQFEE86 pKa = 4.31QSFTDD91 pKa = 5.16FFTQTSNEE99 pKa = 4.05FVNHH103 pKa = 5.77YY104 pKa = 10.29EE105 pKa = 4.01SDD107 pKa = 3.91NII109 pKa = 4.19
Molecular weight: 12.59 kDa
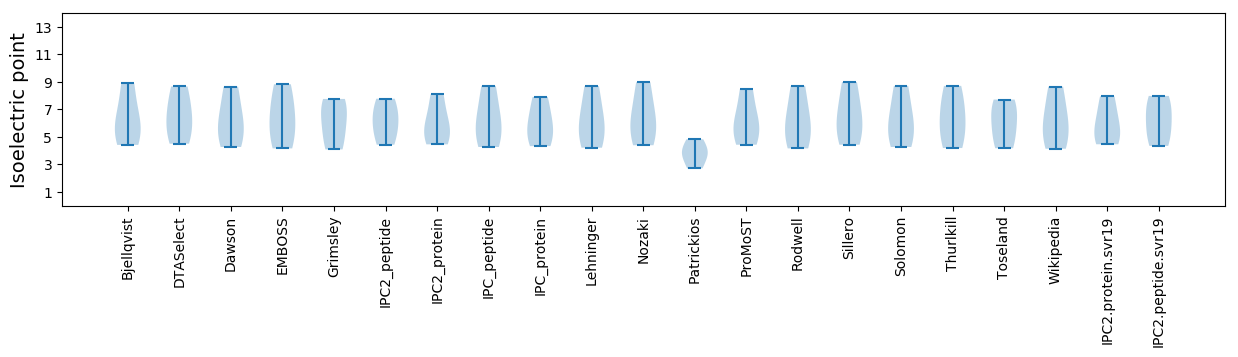
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1FVT1|A0A4V1FVT1_9VIRU Replication initiator protein OS=Capybara microvirus Cap3_SP_416 OX=2585448 PE=4 SV=1
MM1 pKa = 7.82CIAPLEE7 pKa = 4.51IINPKK12 pKa = 9.91LDD14 pKa = 3.67PRR16 pKa = 11.84STDD19 pKa = 3.21PTLLEE24 pKa = 4.37VGCGKK29 pKa = 10.51CEE31 pKa = 3.68EE32 pKa = 4.8CRR34 pKa = 11.84NQRR37 pKa = 11.84KK38 pKa = 9.98DD39 pKa = 2.88EE40 pKa = 4.6WIVRR44 pKa = 11.84TYY46 pKa = 11.08FQMLHH51 pKa = 4.95NRR53 pKa = 11.84EE54 pKa = 3.61RR55 pKa = 11.84GYY57 pKa = 11.54YY58 pKa = 8.32NTFFTLTYY66 pKa = 10.79SDD68 pKa = 4.51DD69 pKa = 4.2CLPHH73 pKa = 8.15LLIDD77 pKa = 4.56GKK79 pKa = 10.27MIPCFNKK86 pKa = 9.53QDD88 pKa = 2.92IFQFFKK94 pKa = 10.49TLKK97 pKa = 8.22KK98 pKa = 10.1HH99 pKa = 5.09LNRR102 pKa = 11.84RR103 pKa = 11.84YY104 pKa = 10.37GINDD108 pKa = 3.18TNYY111 pKa = 10.83LVGCEE116 pKa = 4.39YY117 pKa = 11.2GDD119 pKa = 3.39HH120 pKa = 5.59TQRR123 pKa = 11.84SHH125 pKa = 3.9YY126 pKa = 10.07HH127 pKa = 6.03IMLGCPEE134 pKa = 5.05HH135 pKa = 6.11GTKK138 pKa = 10.31EE139 pKa = 4.06DD140 pKa = 4.18LDD142 pKa = 4.15PRR144 pKa = 11.84DD145 pKa = 3.97LFQFCKK151 pKa = 10.65NYY153 pKa = 8.95WKK155 pKa = 10.46HH156 pKa = 6.63GFMFPRR162 pKa = 11.84VFEE165 pKa = 4.27GGRR168 pKa = 11.84DD169 pKa = 3.42SHH171 pKa = 7.7GYY173 pKa = 7.64DD174 pKa = 3.19HH175 pKa = 7.43KK176 pKa = 10.96PFLVTGDD183 pKa = 3.46ILAPAQYY190 pKa = 9.87ISKK193 pKa = 9.77YY194 pKa = 7.76VCKK197 pKa = 10.57DD198 pKa = 2.99LAFYY202 pKa = 9.55DD203 pKa = 3.56IPEE206 pKa = 4.26ISSLSDD212 pKa = 3.1RR213 pKa = 11.84LDD215 pKa = 3.36KK216 pKa = 11.7EE217 pKa = 4.03EE218 pKa = 3.87FKK220 pKa = 10.78KK221 pKa = 10.94IKK223 pKa = 10.33GSLPFLTVSRR233 pKa = 11.84GFGLYY238 pKa = 10.47LRR240 pKa = 11.84DD241 pKa = 3.84VIMKK245 pKa = 10.16SDD247 pKa = 3.21LHH249 pKa = 7.74NIFKK253 pKa = 10.9YY254 pKa = 10.18GICFEE259 pKa = 4.6GNPRR263 pKa = 11.84RR264 pKa = 11.84HH265 pKa = 6.71KK266 pKa = 10.18IPPYY270 pKa = 9.71ILRR273 pKa = 11.84KK274 pKa = 7.84MCYY277 pKa = 9.93DD278 pKa = 2.95IDD280 pKa = 3.86KK281 pKa = 10.47RR282 pKa = 11.84VVDD285 pKa = 4.6GEE287 pKa = 4.53SKK289 pKa = 10.97VYY291 pKa = 9.67WKK293 pKa = 10.87LNEE296 pKa = 3.85FGKK299 pKa = 10.27QIRR302 pKa = 11.84LQNLDD307 pKa = 3.88NIINSTYY314 pKa = 8.97TSLYY318 pKa = 10.63KK319 pKa = 10.54EE320 pKa = 4.12FSFINLNSDD329 pKa = 3.59YY330 pKa = 9.96MRR332 pKa = 11.84KK333 pKa = 9.19FLSEE337 pKa = 3.83YY338 pKa = 10.25GYY340 pKa = 9.56TSSSFRR346 pKa = 11.84KK347 pKa = 10.51YY348 pKa = 10.39IDD350 pKa = 4.17DD351 pKa = 4.45LLDD354 pKa = 3.61GRR356 pKa = 11.84SFKK359 pKa = 11.05DD360 pKa = 3.16LAIYY364 pKa = 10.02RR365 pKa = 11.84YY366 pKa = 9.78LYY368 pKa = 10.34RR369 pKa = 11.84NRR371 pKa = 11.84ICLHH375 pKa = 5.68EE376 pKa = 5.45SGMVIEE382 pKa = 5.16PGFVSPDD389 pKa = 2.98SSLTEE394 pKa = 3.6KK395 pKa = 10.4RR396 pKa = 11.84VYY398 pKa = 10.18TRR400 pKa = 11.84FFKK403 pKa = 9.87HH404 pKa = 5.06TLYY407 pKa = 10.85NSSIPVMDD415 pKa = 4.09IFRR418 pKa = 11.84DD419 pKa = 3.73HH420 pKa = 7.15RR421 pKa = 11.84GEE423 pKa = 4.15KK424 pKa = 9.93YY425 pKa = 10.26KK426 pKa = 10.99SSSFSKK432 pKa = 10.66KK433 pKa = 10.42FIQDD437 pKa = 3.76TCCFTYY443 pKa = 10.86NSLPLFSGFDD453 pKa = 5.41LILDD457 pKa = 4.05WYY459 pKa = 10.31KK460 pKa = 11.09RR461 pKa = 11.84MSIFHH466 pKa = 6.54KK467 pKa = 10.28KK468 pKa = 9.79YY469 pKa = 9.39NLEE472 pKa = 3.97HH473 pKa = 7.0RR474 pKa = 11.84SIQYY478 pKa = 9.85RR479 pKa = 11.84YY480 pKa = 10.37YY481 pKa = 11.17NYY483 pKa = 9.26IHH485 pKa = 6.25QKK487 pKa = 9.85FVEE490 pKa = 4.44HH491 pKa = 6.57VPP493 pKa = 3.45
MM1 pKa = 7.82CIAPLEE7 pKa = 4.51IINPKK12 pKa = 9.91LDD14 pKa = 3.67PRR16 pKa = 11.84STDD19 pKa = 3.21PTLLEE24 pKa = 4.37VGCGKK29 pKa = 10.51CEE31 pKa = 3.68EE32 pKa = 4.8CRR34 pKa = 11.84NQRR37 pKa = 11.84KK38 pKa = 9.98DD39 pKa = 2.88EE40 pKa = 4.6WIVRR44 pKa = 11.84TYY46 pKa = 11.08FQMLHH51 pKa = 4.95NRR53 pKa = 11.84EE54 pKa = 3.61RR55 pKa = 11.84GYY57 pKa = 11.54YY58 pKa = 8.32NTFFTLTYY66 pKa = 10.79SDD68 pKa = 4.51DD69 pKa = 4.2CLPHH73 pKa = 8.15LLIDD77 pKa = 4.56GKK79 pKa = 10.27MIPCFNKK86 pKa = 9.53QDD88 pKa = 2.92IFQFFKK94 pKa = 10.49TLKK97 pKa = 8.22KK98 pKa = 10.1HH99 pKa = 5.09LNRR102 pKa = 11.84RR103 pKa = 11.84YY104 pKa = 10.37GINDD108 pKa = 3.18TNYY111 pKa = 10.83LVGCEE116 pKa = 4.39YY117 pKa = 11.2GDD119 pKa = 3.39HH120 pKa = 5.59TQRR123 pKa = 11.84SHH125 pKa = 3.9YY126 pKa = 10.07HH127 pKa = 6.03IMLGCPEE134 pKa = 5.05HH135 pKa = 6.11GTKK138 pKa = 10.31EE139 pKa = 4.06DD140 pKa = 4.18LDD142 pKa = 4.15PRR144 pKa = 11.84DD145 pKa = 3.97LFQFCKK151 pKa = 10.65NYY153 pKa = 8.95WKK155 pKa = 10.46HH156 pKa = 6.63GFMFPRR162 pKa = 11.84VFEE165 pKa = 4.27GGRR168 pKa = 11.84DD169 pKa = 3.42SHH171 pKa = 7.7GYY173 pKa = 7.64DD174 pKa = 3.19HH175 pKa = 7.43KK176 pKa = 10.96PFLVTGDD183 pKa = 3.46ILAPAQYY190 pKa = 9.87ISKK193 pKa = 9.77YY194 pKa = 7.76VCKK197 pKa = 10.57DD198 pKa = 2.99LAFYY202 pKa = 9.55DD203 pKa = 3.56IPEE206 pKa = 4.26ISSLSDD212 pKa = 3.1RR213 pKa = 11.84LDD215 pKa = 3.36KK216 pKa = 11.7EE217 pKa = 4.03EE218 pKa = 3.87FKK220 pKa = 10.78KK221 pKa = 10.94IKK223 pKa = 10.33GSLPFLTVSRR233 pKa = 11.84GFGLYY238 pKa = 10.47LRR240 pKa = 11.84DD241 pKa = 3.84VIMKK245 pKa = 10.16SDD247 pKa = 3.21LHH249 pKa = 7.74NIFKK253 pKa = 10.9YY254 pKa = 10.18GICFEE259 pKa = 4.6GNPRR263 pKa = 11.84RR264 pKa = 11.84HH265 pKa = 6.71KK266 pKa = 10.18IPPYY270 pKa = 9.71ILRR273 pKa = 11.84KK274 pKa = 7.84MCYY277 pKa = 9.93DD278 pKa = 2.95IDD280 pKa = 3.86KK281 pKa = 10.47RR282 pKa = 11.84VVDD285 pKa = 4.6GEE287 pKa = 4.53SKK289 pKa = 10.97VYY291 pKa = 9.67WKK293 pKa = 10.87LNEE296 pKa = 3.85FGKK299 pKa = 10.27QIRR302 pKa = 11.84LQNLDD307 pKa = 3.88NIINSTYY314 pKa = 8.97TSLYY318 pKa = 10.63KK319 pKa = 10.54EE320 pKa = 4.12FSFINLNSDD329 pKa = 3.59YY330 pKa = 9.96MRR332 pKa = 11.84KK333 pKa = 9.19FLSEE337 pKa = 3.83YY338 pKa = 10.25GYY340 pKa = 9.56TSSSFRR346 pKa = 11.84KK347 pKa = 10.51YY348 pKa = 10.39IDD350 pKa = 4.17DD351 pKa = 4.45LLDD354 pKa = 3.61GRR356 pKa = 11.84SFKK359 pKa = 11.05DD360 pKa = 3.16LAIYY364 pKa = 10.02RR365 pKa = 11.84YY366 pKa = 9.78LYY368 pKa = 10.34RR369 pKa = 11.84NRR371 pKa = 11.84ICLHH375 pKa = 5.68EE376 pKa = 5.45SGMVIEE382 pKa = 5.16PGFVSPDD389 pKa = 2.98SSLTEE394 pKa = 3.6KK395 pKa = 10.4RR396 pKa = 11.84VYY398 pKa = 10.18TRR400 pKa = 11.84FFKK403 pKa = 9.87HH404 pKa = 5.06TLYY407 pKa = 10.85NSSIPVMDD415 pKa = 4.09IFRR418 pKa = 11.84DD419 pKa = 3.73HH420 pKa = 7.15RR421 pKa = 11.84GEE423 pKa = 4.15KK424 pKa = 9.93YY425 pKa = 10.26KK426 pKa = 10.99SSSFSKK432 pKa = 10.66KK433 pKa = 10.42FIQDD437 pKa = 3.76TCCFTYY443 pKa = 10.86NSLPLFSGFDD453 pKa = 5.41LILDD457 pKa = 4.05WYY459 pKa = 10.31KK460 pKa = 11.09RR461 pKa = 11.84MSIFHH466 pKa = 6.54KK467 pKa = 10.28KK468 pKa = 9.79YY469 pKa = 9.39NLEE472 pKa = 3.97HH473 pKa = 7.0RR474 pKa = 11.84SIQYY478 pKa = 9.85RR479 pKa = 11.84YY480 pKa = 10.37YY481 pKa = 11.17NYY483 pKa = 9.26IHH485 pKa = 6.25QKK487 pKa = 9.85FVEE490 pKa = 4.44HH491 pKa = 6.57VPP493 pKa = 3.45
Molecular weight: 58.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1611 |
109 |
661 |
402.8 |
46.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.593 ± 1.737 | 1.614 ± 0.577 |
6.083 ± 0.456 | 5.276 ± 0.632 |
6.269 ± 0.71 | 5.028 ± 0.477 |
2.607 ± 0.514 | 6.021 ± 0.65 |
5.897 ± 1.033 | 9.497 ± 0.352 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.986 ± 0.24 | 6.642 ± 0.823 |
4.345 ± 0.47 | 4.904 ± 1.061 |
4.593 ± 0.855 | 9.249 ± 0.826 |
4.531 ± 0.395 | 4.718 ± 0.823 |
0.869 ± 0.139 | 5.276 ± 1.087 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
