
Leptospira phage vB_LalZ_80412-LE1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; unclassified Myoviridae
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
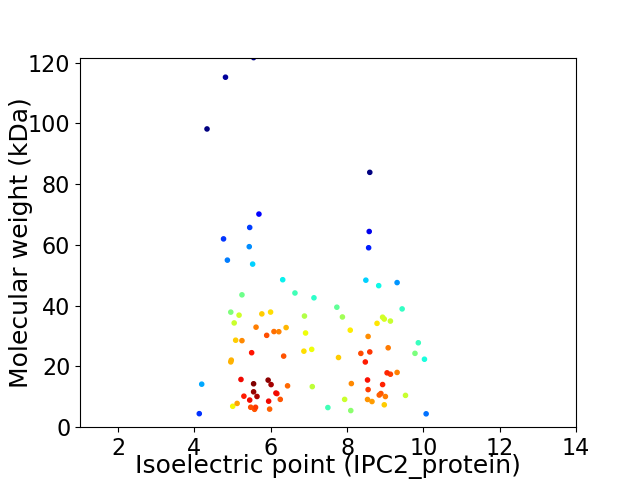
Virtual 2D-PAGE plot for 96 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S5WIH2|S5WIH2_9CAUD Uncharacterized protein OS=Leptospira phage vB_LalZ_80412-LE1 OX=1334242 GN=LEP1GSC193_0783 PE=4 SV=1
MM1 pKa = 7.9KK2 pKa = 10.29LRR4 pKa = 11.84FVGIVFFGLSILFILNFSLLNGISNIFNAVASAIAGGIPQKK45 pKa = 10.97LPIIQAGPDD54 pKa = 3.68GSASTQVEE62 pKa = 4.05IEE64 pKa = 4.09IPPGKK69 pKa = 10.21VIPEE73 pKa = 4.43LSLSYY78 pKa = 10.88NSNGGNGVVGYY89 pKa = 9.41GWSLNGVPTISRR101 pKa = 11.84NPSTGINYY109 pKa = 9.8NGSDD113 pKa = 3.7SYY115 pKa = 11.59ISGLAGEE122 pKa = 5.08LLDD125 pKa = 4.73ISGNKK130 pKa = 7.71TKK132 pKa = 10.71YY133 pKa = 9.68HH134 pKa = 5.88SKK136 pKa = 10.12KK137 pKa = 10.16EE138 pKa = 3.94SWVLYY143 pKa = 9.44EE144 pKa = 4.12PQGTCGDD151 pKa = 4.71GPCTWIATDD160 pKa = 3.61KK161 pKa = 10.89DD162 pKa = 3.35GKK164 pKa = 10.69RR165 pKa = 11.84FIFGGSIDD173 pKa = 3.6SRR175 pKa = 11.84IPALGRR181 pKa = 11.84TAGSIRR187 pKa = 11.84EE188 pKa = 4.0WALSRR193 pKa = 11.84EE194 pKa = 4.32EE195 pKa = 4.77DD196 pKa = 3.7SHH198 pKa = 8.19GNGYY202 pKa = 10.48DD203 pKa = 3.11ITYY206 pKa = 9.63TPIDD210 pKa = 3.94VTNGDD215 pKa = 4.35YY216 pKa = 11.13YY217 pKa = 10.67PNTITYY223 pKa = 9.68NDD225 pKa = 3.21RR226 pKa = 11.84TIRR229 pKa = 11.84FNYY232 pKa = 9.07EE233 pKa = 3.22NRR235 pKa = 11.84NDD237 pKa = 4.19KK238 pKa = 10.51IPNYY242 pKa = 10.7SLGTLIRR249 pKa = 11.84TQRR252 pKa = 11.84RR253 pKa = 11.84LNTIDD258 pKa = 3.48VLVGGTTFRR267 pKa = 11.84TYY269 pKa = 11.37DD270 pKa = 3.47LDD272 pKa = 3.95YY273 pKa = 8.67TTGPVTGRR281 pKa = 11.84SVLQKK286 pKa = 10.17IKK288 pKa = 10.59RR289 pKa = 11.84SGSNTFGSEE298 pKa = 3.43NFADD302 pKa = 5.02LNFTYY307 pKa = 10.12TNHH310 pKa = 6.55SGNFSPGNIDD320 pKa = 3.71YY321 pKa = 7.53QTLSNTISMAVFMPNIALDD340 pKa = 3.41ILNVYY345 pKa = 9.93FNGGLPYY352 pKa = 10.31HH353 pKa = 6.91PSALDD358 pKa = 3.7SNVDD362 pKa = 2.99ASLQYY367 pKa = 9.96IVKK370 pKa = 10.1VPVPDD375 pKa = 4.69RR376 pKa = 11.84NACNLGFASCLCAALPLCWGGNAGFFNYY404 pKa = 10.29LAGNCLSFLSWGGPGACDD422 pKa = 3.46NGVDD426 pKa = 3.95SALTAWLPMDD436 pKa = 4.78LNGDD440 pKa = 4.43GILDD444 pKa = 4.25FATLNGNEE452 pKa = 4.42TNGSIHH458 pKa = 6.7LVGHH462 pKa = 5.9IQRR465 pKa = 11.84IGQGPVTFNGPNIPIHH481 pKa = 6.44YY482 pKa = 7.24NTYY485 pKa = 8.81YY486 pKa = 10.7QPVDD490 pKa = 3.84LNGDD494 pKa = 3.74GKK496 pKa = 9.39TDD498 pKa = 3.86FAYY501 pKa = 10.26EE502 pKa = 4.65DD503 pKa = 4.67GGQLWAVYY511 pKa = 8.77STGGNFSSPVAFGNVSLAGSNRR533 pKa = 11.84DD534 pKa = 3.13MKK536 pKa = 11.09VFSPYY541 pKa = 10.17EE542 pKa = 3.65YY543 pKa = 10.09RR544 pKa = 11.84FQYY547 pKa = 10.57SPSNTTPLASNRR559 pKa = 11.84ATSDD563 pKa = 3.14FFADD567 pKa = 3.7MNGDD571 pKa = 3.89DD572 pKa = 4.41LTDD575 pKa = 4.17FVHH578 pKa = 7.09SNGGSFSIYY587 pKa = 9.98INRR590 pKa = 11.84EE591 pKa = 3.86TYY593 pKa = 9.82FDD595 pKa = 3.72NPIVIGGGNDD605 pKa = 3.55FYY607 pKa = 10.78INSMIDD613 pKa = 2.97MTGDD617 pKa = 3.26GKK619 pKa = 11.11SDD621 pKa = 3.68YY622 pKa = 10.27VQLIATYY629 pKa = 10.67DD630 pKa = 3.57NSTLTTLQAQKK641 pKa = 10.94AALDD645 pKa = 3.64VLMAQYY651 pKa = 10.47QVDD654 pKa = 3.82HH655 pKa = 6.74ARR657 pKa = 11.84VKK659 pKa = 10.68AVADD663 pKa = 4.7LYY665 pKa = 10.64PLALGVTVDD674 pKa = 3.69ANEE677 pKa = 3.86FQYY680 pKa = 10.75MIDD683 pKa = 3.63YY684 pKa = 10.85LNTNGYY690 pKa = 10.35DD691 pKa = 3.5PLADD695 pKa = 3.76FWEE698 pKa = 5.04SNGSGYY704 pKa = 10.94AYY706 pKa = 10.45NSSEE710 pKa = 3.93IGNLQTSLEE719 pKa = 4.43NIVSAKK725 pKa = 9.46MNFVGQQSQAINIQIASIYY744 pKa = 9.98AQGTLGQATYY754 pKa = 10.74ALQVRR759 pKa = 11.84TFNTSNGTSQVQTFPISNSIDD780 pKa = 3.37ADD782 pKa = 3.81RR783 pKa = 11.84STLADD788 pKa = 3.54VNGDD792 pKa = 3.42GALDD796 pKa = 3.72FVSFIGTQVSVSIFTGNGFAPPVVTGLNAGNGKK829 pKa = 9.05NLVQFNFGEE838 pKa = 4.4VNGDD842 pKa = 3.68GLSDD846 pKa = 3.61LVLFNRR852 pKa = 11.84EE853 pKa = 3.49SHH855 pKa = 7.01IIEE858 pKa = 4.71TYY860 pKa = 10.57LSRR863 pKa = 11.84GDD865 pKa = 3.69GGFTLSGGYY874 pKa = 9.94SFGGFSTQEE883 pKa = 3.87YY884 pKa = 8.53TDD886 pKa = 3.44SNGIEE891 pKa = 4.19RR892 pKa = 11.84SDD894 pKa = 4.02LYY896 pKa = 10.99QISLQDD902 pKa = 3.33INLDD906 pKa = 4.24GISS909 pKa = 3.31
MM1 pKa = 7.9KK2 pKa = 10.29LRR4 pKa = 11.84FVGIVFFGLSILFILNFSLLNGISNIFNAVASAIAGGIPQKK45 pKa = 10.97LPIIQAGPDD54 pKa = 3.68GSASTQVEE62 pKa = 4.05IEE64 pKa = 4.09IPPGKK69 pKa = 10.21VIPEE73 pKa = 4.43LSLSYY78 pKa = 10.88NSNGGNGVVGYY89 pKa = 9.41GWSLNGVPTISRR101 pKa = 11.84NPSTGINYY109 pKa = 9.8NGSDD113 pKa = 3.7SYY115 pKa = 11.59ISGLAGEE122 pKa = 5.08LLDD125 pKa = 4.73ISGNKK130 pKa = 7.71TKK132 pKa = 10.71YY133 pKa = 9.68HH134 pKa = 5.88SKK136 pKa = 10.12KK137 pKa = 10.16EE138 pKa = 3.94SWVLYY143 pKa = 9.44EE144 pKa = 4.12PQGTCGDD151 pKa = 4.71GPCTWIATDD160 pKa = 3.61KK161 pKa = 10.89DD162 pKa = 3.35GKK164 pKa = 10.69RR165 pKa = 11.84FIFGGSIDD173 pKa = 3.6SRR175 pKa = 11.84IPALGRR181 pKa = 11.84TAGSIRR187 pKa = 11.84EE188 pKa = 4.0WALSRR193 pKa = 11.84EE194 pKa = 4.32EE195 pKa = 4.77DD196 pKa = 3.7SHH198 pKa = 8.19GNGYY202 pKa = 10.48DD203 pKa = 3.11ITYY206 pKa = 9.63TPIDD210 pKa = 3.94VTNGDD215 pKa = 4.35YY216 pKa = 11.13YY217 pKa = 10.67PNTITYY223 pKa = 9.68NDD225 pKa = 3.21RR226 pKa = 11.84TIRR229 pKa = 11.84FNYY232 pKa = 9.07EE233 pKa = 3.22NRR235 pKa = 11.84NDD237 pKa = 4.19KK238 pKa = 10.51IPNYY242 pKa = 10.7SLGTLIRR249 pKa = 11.84TQRR252 pKa = 11.84RR253 pKa = 11.84LNTIDD258 pKa = 3.48VLVGGTTFRR267 pKa = 11.84TYY269 pKa = 11.37DD270 pKa = 3.47LDD272 pKa = 3.95YY273 pKa = 8.67TTGPVTGRR281 pKa = 11.84SVLQKK286 pKa = 10.17IKK288 pKa = 10.59RR289 pKa = 11.84SGSNTFGSEE298 pKa = 3.43NFADD302 pKa = 5.02LNFTYY307 pKa = 10.12TNHH310 pKa = 6.55SGNFSPGNIDD320 pKa = 3.71YY321 pKa = 7.53QTLSNTISMAVFMPNIALDD340 pKa = 3.41ILNVYY345 pKa = 9.93FNGGLPYY352 pKa = 10.31HH353 pKa = 6.91PSALDD358 pKa = 3.7SNVDD362 pKa = 2.99ASLQYY367 pKa = 9.96IVKK370 pKa = 10.1VPVPDD375 pKa = 4.69RR376 pKa = 11.84NACNLGFASCLCAALPLCWGGNAGFFNYY404 pKa = 10.29LAGNCLSFLSWGGPGACDD422 pKa = 3.46NGVDD426 pKa = 3.95SALTAWLPMDD436 pKa = 4.78LNGDD440 pKa = 4.43GILDD444 pKa = 4.25FATLNGNEE452 pKa = 4.42TNGSIHH458 pKa = 6.7LVGHH462 pKa = 5.9IQRR465 pKa = 11.84IGQGPVTFNGPNIPIHH481 pKa = 6.44YY482 pKa = 7.24NTYY485 pKa = 8.81YY486 pKa = 10.7QPVDD490 pKa = 3.84LNGDD494 pKa = 3.74GKK496 pKa = 9.39TDD498 pKa = 3.86FAYY501 pKa = 10.26EE502 pKa = 4.65DD503 pKa = 4.67GGQLWAVYY511 pKa = 8.77STGGNFSSPVAFGNVSLAGSNRR533 pKa = 11.84DD534 pKa = 3.13MKK536 pKa = 11.09VFSPYY541 pKa = 10.17EE542 pKa = 3.65YY543 pKa = 10.09RR544 pKa = 11.84FQYY547 pKa = 10.57SPSNTTPLASNRR559 pKa = 11.84ATSDD563 pKa = 3.14FFADD567 pKa = 3.7MNGDD571 pKa = 3.89DD572 pKa = 4.41LTDD575 pKa = 4.17FVHH578 pKa = 7.09SNGGSFSIYY587 pKa = 9.98INRR590 pKa = 11.84EE591 pKa = 3.86TYY593 pKa = 9.82FDD595 pKa = 3.72NPIVIGGGNDD605 pKa = 3.55FYY607 pKa = 10.78INSMIDD613 pKa = 2.97MTGDD617 pKa = 3.26GKK619 pKa = 11.11SDD621 pKa = 3.68YY622 pKa = 10.27VQLIATYY629 pKa = 10.67DD630 pKa = 3.57NSTLTTLQAQKK641 pKa = 10.94AALDD645 pKa = 3.64VLMAQYY651 pKa = 10.47QVDD654 pKa = 3.82HH655 pKa = 6.74ARR657 pKa = 11.84VKK659 pKa = 10.68AVADD663 pKa = 4.7LYY665 pKa = 10.64PLALGVTVDD674 pKa = 3.69ANEE677 pKa = 3.86FQYY680 pKa = 10.75MIDD683 pKa = 3.63YY684 pKa = 10.85LNTNGYY690 pKa = 10.35DD691 pKa = 3.5PLADD695 pKa = 3.76FWEE698 pKa = 5.04SNGSGYY704 pKa = 10.94AYY706 pKa = 10.45NSSEE710 pKa = 3.93IGNLQTSLEE719 pKa = 4.43NIVSAKK725 pKa = 9.46MNFVGQQSQAINIQIASIYY744 pKa = 9.98AQGTLGQATYY754 pKa = 10.74ALQVRR759 pKa = 11.84TFNTSNGTSQVQTFPISNSIDD780 pKa = 3.37ADD782 pKa = 3.81RR783 pKa = 11.84STLADD788 pKa = 3.54VNGDD792 pKa = 3.42GALDD796 pKa = 3.72FVSFIGTQVSVSIFTGNGFAPPVVTGLNAGNGKK829 pKa = 9.05NLVQFNFGEE838 pKa = 4.4VNGDD842 pKa = 3.68GLSDD846 pKa = 3.61LVLFNRR852 pKa = 11.84EE853 pKa = 3.49SHH855 pKa = 7.01IIEE858 pKa = 4.71TYY860 pKa = 10.57LSRR863 pKa = 11.84GDD865 pKa = 3.69GGFTLSGGYY874 pKa = 9.94SFGGFSTQEE883 pKa = 3.87YY884 pKa = 8.53TDD886 pKa = 3.44SNGIEE891 pKa = 4.19RR892 pKa = 11.84SDD894 pKa = 4.02LYY896 pKa = 10.99QISLQDD902 pKa = 3.33INLDD906 pKa = 4.24GISS909 pKa = 3.31
Molecular weight: 98.14 kDa
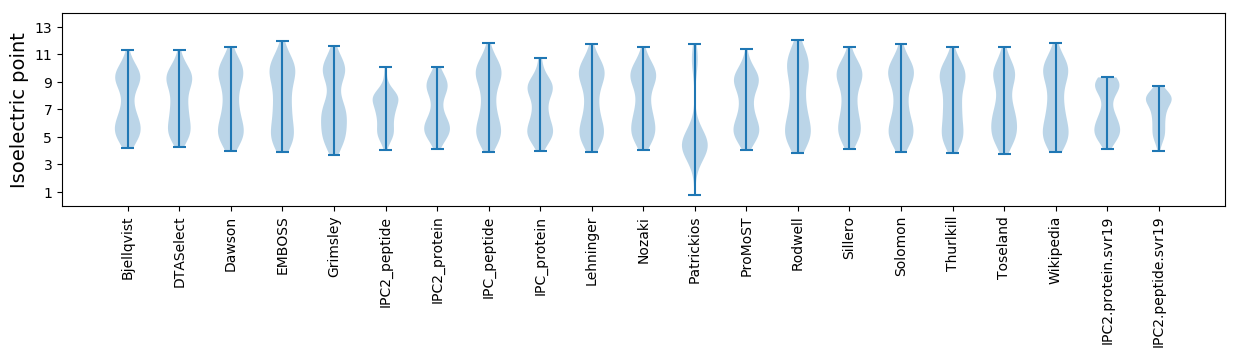
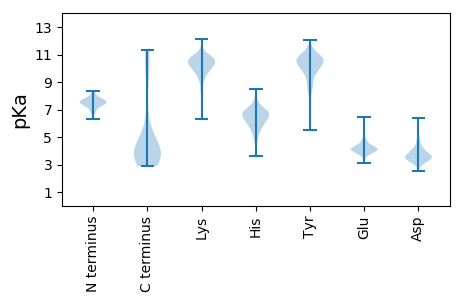
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5VXX3|S5VXX3_9CAUD Uncharacterized protein OS=Leptospira phage vB_LalZ_80412-LE1 OX=1334242 GN=LEP1GSC193_0717 PE=4 SV=1
MM1 pKa = 7.63CSSKK5 pKa = 11.32SNAFNGGSSTSSPVLLLTQLIKK27 pKa = 10.5FAITLKK33 pKa = 10.36SLCFSSLLGPYY44 pKa = 8.87PQARR48 pKa = 11.84LYY50 pKa = 10.88SSSSKK55 pKa = 10.31KK56 pKa = 10.32VCLEE60 pKa = 3.67SPIFIFYY67 pKa = 10.3RR68 pKa = 11.84FSIRR72 pKa = 11.84SIGASSFSRR81 pKa = 11.84ILGCIVRR88 pKa = 11.84RR89 pKa = 11.84YY90 pKa = 9.65RR91 pKa = 11.84IVYY94 pKa = 9.04GILVRR99 pKa = 11.84ILSLTFVQNLGLMFLFKK116 pKa = 10.88LIGSTSQIFHH126 pKa = 7.28PKK128 pKa = 9.99AKK130 pKa = 9.88PLVSLFEE137 pKa = 3.94VMLIYY142 pKa = 10.56FLLPKK147 pKa = 10.02PVANSQFFPITQSLRR162 pKa = 11.84ILHH165 pKa = 6.06NSEE168 pKa = 3.91SRR170 pKa = 11.84NPHH173 pKa = 6.96PDD175 pKa = 2.89LQRR178 pKa = 11.84YY179 pKa = 8.0LLQILPFGLSTILFRR194 pKa = 11.84NSRR197 pKa = 11.84NN198 pKa = 3.32
MM1 pKa = 7.63CSSKK5 pKa = 11.32SNAFNGGSSTSSPVLLLTQLIKK27 pKa = 10.5FAITLKK33 pKa = 10.36SLCFSSLLGPYY44 pKa = 8.87PQARR48 pKa = 11.84LYY50 pKa = 10.88SSSSKK55 pKa = 10.31KK56 pKa = 10.32VCLEE60 pKa = 3.67SPIFIFYY67 pKa = 10.3RR68 pKa = 11.84FSIRR72 pKa = 11.84SIGASSFSRR81 pKa = 11.84ILGCIVRR88 pKa = 11.84RR89 pKa = 11.84YY90 pKa = 9.65RR91 pKa = 11.84IVYY94 pKa = 9.04GILVRR99 pKa = 11.84ILSLTFVQNLGLMFLFKK116 pKa = 10.88LIGSTSQIFHH126 pKa = 7.28PKK128 pKa = 9.99AKK130 pKa = 9.88PLVSLFEE137 pKa = 3.94VMLIYY142 pKa = 10.56FLLPKK147 pKa = 10.02PVANSQFFPITQSLRR162 pKa = 11.84ILHH165 pKa = 6.06NSEE168 pKa = 3.91SRR170 pKa = 11.84NPHH173 pKa = 6.96PDD175 pKa = 2.89LQRR178 pKa = 11.84YY179 pKa = 8.0LLQILPFGLSTILFRR194 pKa = 11.84NSRR197 pKa = 11.84NN198 pKa = 3.32
Molecular weight: 22.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
24630 |
37 |
1116 |
256.6 |
28.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.876 ± 0.232 | 0.824 ± 0.072 |
4.815 ± 0.169 | 6.764 ± 0.414 |
5.493 ± 0.246 | 7.077 ± 0.393 |
1.429 ± 0.082 | 7.089 ± 0.237 |
7.84 ± 0.536 | 9.704 ± 0.259 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.433 ± 0.089 | 6.074 ± 0.193 |
4.056 ± 0.166 | 3.228 ± 0.114 |
4.499 ± 0.169 | 8.368 ± 0.283 |
6.098 ± 0.291 | 5.416 ± 0.165 |
1.137 ± 0.07 | 3.78 ± 0.179 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
