
Vibrio sp. 10N.286.49.B3
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Vibrio; unclassified Vibrio
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
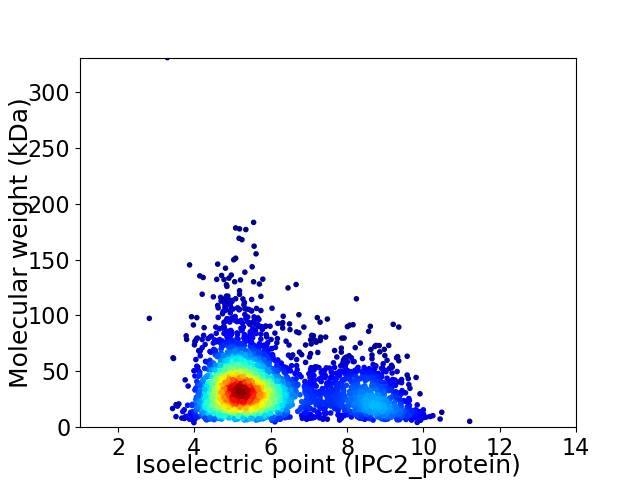
Virtual 2D-PAGE plot for 3192 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N7DBZ1|A0A2N7DBZ1_9VIBR 2-dehydro-3-deoxyphosphooctonate aldolase OS=Vibrio sp. 10N.286.49.B3 OX=1880855 GN=kdsA PE=3 SV=1
MM1 pKa = 7.78EE2 pKa = 4.55KK3 pKa = 9.63TFKK6 pKa = 10.85LSLVFSAILLAGCGDD21 pKa = 3.88DD22 pKa = 4.38TEE24 pKa = 5.49SSGAPTTPVYY34 pKa = 10.98EE35 pKa = 4.83DD36 pKa = 5.2FIQEE40 pKa = 4.18SLEE43 pKa = 4.06QPTTIQFTLSGQNAAVPLPSFALLDD68 pKa = 3.91TSDD71 pKa = 3.8GTLGLPTGGDD81 pKa = 3.62DD82 pKa = 4.74ALSNPIAAMNTADD95 pKa = 4.06GFSTTMPISLTFEE108 pKa = 4.31GGDD111 pKa = 3.36FGEE114 pKa = 4.54TQNILTSGIYY124 pKa = 9.79VVEE127 pKa = 4.46LSDD130 pKa = 4.42GLTGSPTPQAILSAPTDD147 pKa = 4.05FVTLAQGSNLTIIFNEE163 pKa = 3.86PLNPKK168 pKa = 9.78SNYY171 pKa = 8.95IFAVTNEE178 pKa = 4.05VLDD181 pKa = 3.88SDD183 pKa = 4.48GEE185 pKa = 4.37AVGMSSSYY193 pKa = 11.04AAAKK197 pKa = 10.36SEE199 pKa = 3.93EE200 pKa = 4.21KK201 pKa = 10.26IYY203 pKa = 10.99TEE205 pKa = 4.14GSLAQVQQVTQGVEE219 pKa = 3.78ALFAATGVDD228 pKa = 3.69KK229 pKa = 10.25EE230 pKa = 4.57TIIYY234 pKa = 9.61SSWFITQSVGDD245 pKa = 4.02SLYY248 pKa = 9.58ATKK251 pKa = 10.71GATATGLASGGLNNVWALDD270 pKa = 3.72SQANPNNVDD279 pKa = 3.14LTYY282 pKa = 10.62AYY284 pKa = 9.72IMSFEE289 pKa = 4.11EE290 pKa = 4.0TATFNDD296 pKa = 3.92ALDD299 pKa = 4.09NDD301 pKa = 4.59SDD303 pKa = 4.0FDD305 pKa = 4.94DD306 pKa = 5.71YY307 pKa = 11.44IGSDD311 pKa = 2.99AKK313 pKa = 11.1AGIKK317 pKa = 10.0ALYY320 pKa = 7.78EE321 pKa = 4.12ASDD324 pKa = 3.92AEE326 pKa = 4.64VNVTQGTVNLPHH338 pKa = 7.33YY339 pKa = 10.2LEE341 pKa = 4.49QDD343 pKa = 3.35LATWNSQPFQSAMPSLAKK361 pKa = 10.35ISAALSDD368 pKa = 4.35EE369 pKa = 4.25NEE371 pKa = 4.11QAHH374 pKa = 6.12VAGQLMAAGITPSLLATSPEE394 pKa = 4.09EE395 pKa = 3.71QLKK398 pKa = 10.89LVGQSLTLSDD408 pKa = 4.58GSALDD413 pKa = 3.56SEE415 pKa = 5.09RR416 pKa = 11.84VITQYY421 pKa = 11.55SPVPQVKK428 pKa = 10.16SLEE431 pKa = 4.04AVEE434 pKa = 5.95FILFTPTNSTATEE447 pKa = 3.66IAIYY451 pKa = 9.12QHH453 pKa = 7.18GITSAKK459 pKa = 8.62EE460 pKa = 3.35NAYY463 pKa = 10.53AFAFNQAQQGIAVLAIDD480 pKa = 5.02LPLHH484 pKa = 5.61GTRR487 pKa = 11.84SLDD490 pKa = 3.32EE491 pKa = 4.73DD492 pKa = 3.73RR493 pKa = 11.84SANVDD498 pKa = 3.23TLAYY502 pKa = 10.82LNLTYY507 pKa = 10.9LPVARR512 pKa = 11.84DD513 pKa = 3.62NLRR516 pKa = 11.84QSALDD521 pKa = 3.63VMGLRR526 pKa = 11.84AALAVSSQAGLLTTTPLSAINPVTSSHH553 pKa = 7.38FIGHH557 pKa = 5.77SLGGIVGVSAVAAANEE573 pKa = 4.33SLGSDD578 pKa = 3.54TADD581 pKa = 2.97ALYY584 pKa = 10.5NFSSMSIQNSGGQIANLLLGSTNFGPLIMHH614 pKa = 7.18NIALSLSIGYY624 pKa = 9.91DD625 pKa = 3.09SYY627 pKa = 11.81VAEE630 pKa = 4.43NCTDD634 pKa = 3.68VIEE637 pKa = 4.61QICFAQFSAQATEE650 pKa = 4.09EE651 pKa = 4.12QQAVMSAGFQQFSYY665 pKa = 10.81AAQTVLDD672 pKa = 4.65TIDD675 pKa = 3.85PFTNASHH682 pKa = 6.8LVDD685 pKa = 4.0NATLIPVLMSQVADD699 pKa = 4.34DD700 pKa = 4.02DD701 pKa = 4.45TVPNSVDD708 pKa = 3.8FAPFAGTEE716 pKa = 3.92PLAAQLSLDD725 pKa = 3.85EE726 pKa = 4.52TSTPGTTGGFVKK738 pKa = 9.94FTDD741 pKa = 3.55SAAGHH746 pKa = 5.37STFVAPKK753 pKa = 9.84SDD755 pKa = 3.27LSDD758 pKa = 3.19IVTHH762 pKa = 6.61ATMQSQQTQFIAPTTTPP779 pKa = 3.18
MM1 pKa = 7.78EE2 pKa = 4.55KK3 pKa = 9.63TFKK6 pKa = 10.85LSLVFSAILLAGCGDD21 pKa = 3.88DD22 pKa = 4.38TEE24 pKa = 5.49SSGAPTTPVYY34 pKa = 10.98EE35 pKa = 4.83DD36 pKa = 5.2FIQEE40 pKa = 4.18SLEE43 pKa = 4.06QPTTIQFTLSGQNAAVPLPSFALLDD68 pKa = 3.91TSDD71 pKa = 3.8GTLGLPTGGDD81 pKa = 3.62DD82 pKa = 4.74ALSNPIAAMNTADD95 pKa = 4.06GFSTTMPISLTFEE108 pKa = 4.31GGDD111 pKa = 3.36FGEE114 pKa = 4.54TQNILTSGIYY124 pKa = 9.79VVEE127 pKa = 4.46LSDD130 pKa = 4.42GLTGSPTPQAILSAPTDD147 pKa = 4.05FVTLAQGSNLTIIFNEE163 pKa = 3.86PLNPKK168 pKa = 9.78SNYY171 pKa = 8.95IFAVTNEE178 pKa = 4.05VLDD181 pKa = 3.88SDD183 pKa = 4.48GEE185 pKa = 4.37AVGMSSSYY193 pKa = 11.04AAAKK197 pKa = 10.36SEE199 pKa = 3.93EE200 pKa = 4.21KK201 pKa = 10.26IYY203 pKa = 10.99TEE205 pKa = 4.14GSLAQVQQVTQGVEE219 pKa = 3.78ALFAATGVDD228 pKa = 3.69KK229 pKa = 10.25EE230 pKa = 4.57TIIYY234 pKa = 9.61SSWFITQSVGDD245 pKa = 4.02SLYY248 pKa = 9.58ATKK251 pKa = 10.71GATATGLASGGLNNVWALDD270 pKa = 3.72SQANPNNVDD279 pKa = 3.14LTYY282 pKa = 10.62AYY284 pKa = 9.72IMSFEE289 pKa = 4.11EE290 pKa = 4.0TATFNDD296 pKa = 3.92ALDD299 pKa = 4.09NDD301 pKa = 4.59SDD303 pKa = 4.0FDD305 pKa = 4.94DD306 pKa = 5.71YY307 pKa = 11.44IGSDD311 pKa = 2.99AKK313 pKa = 11.1AGIKK317 pKa = 10.0ALYY320 pKa = 7.78EE321 pKa = 4.12ASDD324 pKa = 3.92AEE326 pKa = 4.64VNVTQGTVNLPHH338 pKa = 7.33YY339 pKa = 10.2LEE341 pKa = 4.49QDD343 pKa = 3.35LATWNSQPFQSAMPSLAKK361 pKa = 10.35ISAALSDD368 pKa = 4.35EE369 pKa = 4.25NEE371 pKa = 4.11QAHH374 pKa = 6.12VAGQLMAAGITPSLLATSPEE394 pKa = 4.09EE395 pKa = 3.71QLKK398 pKa = 10.89LVGQSLTLSDD408 pKa = 4.58GSALDD413 pKa = 3.56SEE415 pKa = 5.09RR416 pKa = 11.84VITQYY421 pKa = 11.55SPVPQVKK428 pKa = 10.16SLEE431 pKa = 4.04AVEE434 pKa = 5.95FILFTPTNSTATEE447 pKa = 3.66IAIYY451 pKa = 9.12QHH453 pKa = 7.18GITSAKK459 pKa = 8.62EE460 pKa = 3.35NAYY463 pKa = 10.53AFAFNQAQQGIAVLAIDD480 pKa = 5.02LPLHH484 pKa = 5.61GTRR487 pKa = 11.84SLDD490 pKa = 3.32EE491 pKa = 4.73DD492 pKa = 3.73RR493 pKa = 11.84SANVDD498 pKa = 3.23TLAYY502 pKa = 10.82LNLTYY507 pKa = 10.9LPVARR512 pKa = 11.84DD513 pKa = 3.62NLRR516 pKa = 11.84QSALDD521 pKa = 3.63VMGLRR526 pKa = 11.84AALAVSSQAGLLTTTPLSAINPVTSSHH553 pKa = 7.38FIGHH557 pKa = 5.77SLGGIVGVSAVAAANEE573 pKa = 4.33SLGSDD578 pKa = 3.54TADD581 pKa = 2.97ALYY584 pKa = 10.5NFSSMSIQNSGGQIANLLLGSTNFGPLIMHH614 pKa = 7.18NIALSLSIGYY624 pKa = 9.91DD625 pKa = 3.09SYY627 pKa = 11.81VAEE630 pKa = 4.43NCTDD634 pKa = 3.68VIEE637 pKa = 4.61QICFAQFSAQATEE650 pKa = 4.09EE651 pKa = 4.12QQAVMSAGFQQFSYY665 pKa = 10.81AAQTVLDD672 pKa = 4.65TIDD675 pKa = 3.85PFTNASHH682 pKa = 6.8LVDD685 pKa = 4.0NATLIPVLMSQVADD699 pKa = 4.34DD700 pKa = 4.02DD701 pKa = 4.45TVPNSVDD708 pKa = 3.8FAPFAGTEE716 pKa = 3.92PLAAQLSLDD725 pKa = 3.85EE726 pKa = 4.52TSTPGTTGGFVKK738 pKa = 9.94FTDD741 pKa = 3.55SAAGHH746 pKa = 5.37STFVAPKK753 pKa = 9.84SDD755 pKa = 3.27LSDD758 pKa = 3.19IVTHH762 pKa = 6.61ATMQSQQTQFIAPTTTPP779 pKa = 3.18
Molecular weight: 81.76 kDa
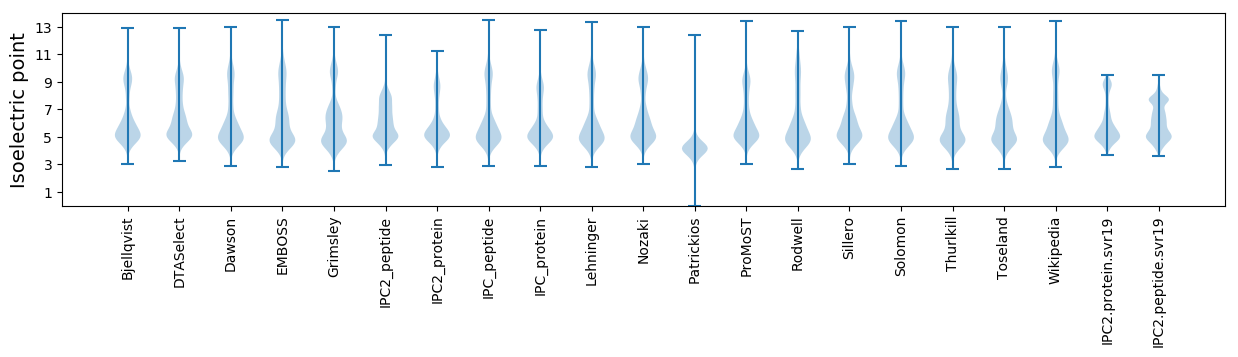
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N7DCD8|A0A2N7DCD8_9VIBR Cyclic pyranopterin monophosphate synthase OS=Vibrio sp. 10N.286.49.B3 OX=1880855 GN=moaC PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 8.99RR4 pKa = 11.84TFQPTVLKK12 pKa = 10.46RR13 pKa = 11.84KK14 pKa = 7.65RR15 pKa = 11.84THH17 pKa = 5.89GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.44NGRR29 pKa = 11.84KK30 pKa = 9.39VINARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.22GRR40 pKa = 11.84ARR42 pKa = 11.84LSKK45 pKa = 10.83
MM1 pKa = 7.69SKK3 pKa = 8.99RR4 pKa = 11.84TFQPTVLKK12 pKa = 10.46RR13 pKa = 11.84KK14 pKa = 7.65RR15 pKa = 11.84THH17 pKa = 5.89GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.44NGRR29 pKa = 11.84KK30 pKa = 9.39VINARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.22GRR40 pKa = 11.84ARR42 pKa = 11.84LSKK45 pKa = 10.83
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1073679 |
35 |
3119 |
336.4 |
37.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.321 ± 0.047 | 1.016 ± 0.014 |
5.694 ± 0.039 | 6.262 ± 0.036 |
4.136 ± 0.03 | 6.691 ± 0.043 |
2.29 ± 0.022 | 6.787 ± 0.033 |
5.322 ± 0.039 | 10.299 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.674 ± 0.025 | 4.432 ± 0.032 |
3.777 ± 0.025 | 4.662 ± 0.036 |
4.325 ± 0.038 | 6.785 ± 0.037 |
5.458 ± 0.032 | 6.799 ± 0.035 |
1.211 ± 0.017 | 3.058 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
