
Maize striate mosaic virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus
Average proteome isoelectric point is 7.51
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
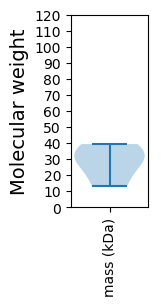
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A286QV14|A0A286QV14_9GEMI Replication-associated protein OS=Maize striate mosaic virus OX=2025388 PE=3 SV=1
MM1 pKa = 7.24SHH3 pKa = 5.59TSFRR7 pKa = 11.84FRR9 pKa = 11.84AKK11 pKa = 10.45NVFLTYY17 pKa = 9.25PRR19 pKa = 11.84CPIGPEE25 pKa = 3.94FLCDD29 pKa = 3.78HH30 pKa = 7.2LWNLVTPYY38 pKa = 10.93DD39 pKa = 3.74PLYY42 pKa = 10.82VHH44 pKa = 6.93VAQEE48 pKa = 3.85NHH50 pKa = 6.79KK51 pKa = 11.0DD52 pKa = 3.66GGLHH56 pKa = 5.08SHH58 pKa = 6.28VLIQTRR64 pKa = 11.84IEE66 pKa = 4.04ISTFDD71 pKa = 3.64PTYY74 pKa = 10.78FDD76 pKa = 3.58YY77 pKa = 10.96TGTSIPGAVVFHH89 pKa = 6.59PNIQACRR96 pKa = 11.84NVRR99 pKa = 11.84DD100 pKa = 3.51CLAYY104 pKa = 9.62IRR106 pKa = 11.84KK107 pKa = 8.28NTINEE112 pKa = 3.86VSKK115 pKa = 11.12GAFKK119 pKa = 10.88CSGAGRR125 pKa = 11.84PKK127 pKa = 10.47KK128 pKa = 9.99QDD130 pKa = 3.29SAPSRR135 pKa = 11.84DD136 pKa = 3.25AKK138 pKa = 9.94MCQIMSSATSRR149 pKa = 11.84SDD151 pKa = 3.35YY152 pKa = 11.09LSMVRR157 pKa = 11.84GAFPFEE163 pKa = 4.23WATKK167 pKa = 9.6LAQFEE172 pKa = 4.47YY173 pKa = 10.32SASKK177 pKa = 10.5LFPDD181 pKa = 4.87APTTNAIPSNIDD193 pKa = 3.2LTCHH197 pKa = 5.53EE198 pKa = 5.77NIMPWLRR205 pKa = 11.84DD206 pKa = 3.36DD207 pKa = 5.24LYY209 pKa = 10.42TVSEE213 pKa = 4.46FSYY216 pKa = 9.75FLCTGSLDD224 pKa = 3.88PKK226 pKa = 11.42GDD228 pKa = 3.92LTWMADD234 pKa = 3.14HH235 pKa = 6.72TRR237 pKa = 11.84NEE239 pKa = 4.23GKK241 pKa = 10.34EE242 pKa = 4.03DD243 pKa = 3.48VPEE246 pKa = 4.39ASTYY250 pKa = 10.43AEE252 pKa = 3.71EE253 pKa = 4.08RR254 pKa = 11.84GRR256 pKa = 11.84ARR258 pKa = 11.84QHH260 pKa = 4.96GQEE263 pKa = 4.18ASGHH267 pKa = 4.62TTTSNTRR274 pKa = 11.84WTTSPGAKK282 pKa = 8.8RR283 pKa = 11.84PRR285 pKa = 11.84MSSS288 pKa = 2.83
MM1 pKa = 7.24SHH3 pKa = 5.59TSFRR7 pKa = 11.84FRR9 pKa = 11.84AKK11 pKa = 10.45NVFLTYY17 pKa = 9.25PRR19 pKa = 11.84CPIGPEE25 pKa = 3.94FLCDD29 pKa = 3.78HH30 pKa = 7.2LWNLVTPYY38 pKa = 10.93DD39 pKa = 3.74PLYY42 pKa = 10.82VHH44 pKa = 6.93VAQEE48 pKa = 3.85NHH50 pKa = 6.79KK51 pKa = 11.0DD52 pKa = 3.66GGLHH56 pKa = 5.08SHH58 pKa = 6.28VLIQTRR64 pKa = 11.84IEE66 pKa = 4.04ISTFDD71 pKa = 3.64PTYY74 pKa = 10.78FDD76 pKa = 3.58YY77 pKa = 10.96TGTSIPGAVVFHH89 pKa = 6.59PNIQACRR96 pKa = 11.84NVRR99 pKa = 11.84DD100 pKa = 3.51CLAYY104 pKa = 9.62IRR106 pKa = 11.84KK107 pKa = 8.28NTINEE112 pKa = 3.86VSKK115 pKa = 11.12GAFKK119 pKa = 10.88CSGAGRR125 pKa = 11.84PKK127 pKa = 10.47KK128 pKa = 9.99QDD130 pKa = 3.29SAPSRR135 pKa = 11.84DD136 pKa = 3.25AKK138 pKa = 9.94MCQIMSSATSRR149 pKa = 11.84SDD151 pKa = 3.35YY152 pKa = 11.09LSMVRR157 pKa = 11.84GAFPFEE163 pKa = 4.23WATKK167 pKa = 9.6LAQFEE172 pKa = 4.47YY173 pKa = 10.32SASKK177 pKa = 10.5LFPDD181 pKa = 4.87APTTNAIPSNIDD193 pKa = 3.2LTCHH197 pKa = 5.53EE198 pKa = 5.77NIMPWLRR205 pKa = 11.84DD206 pKa = 3.36DD207 pKa = 5.24LYY209 pKa = 10.42TVSEE213 pKa = 4.46FSYY216 pKa = 9.75FLCTGSLDD224 pKa = 3.88PKK226 pKa = 11.42GDD228 pKa = 3.92LTWMADD234 pKa = 3.14HH235 pKa = 6.72TRR237 pKa = 11.84NEE239 pKa = 4.23GKK241 pKa = 10.34EE242 pKa = 4.03DD243 pKa = 3.48VPEE246 pKa = 4.39ASTYY250 pKa = 10.43AEE252 pKa = 3.71EE253 pKa = 4.08RR254 pKa = 11.84GRR256 pKa = 11.84ARR258 pKa = 11.84QHH260 pKa = 4.96GQEE263 pKa = 4.18ASGHH267 pKa = 4.62TTTSNTRR274 pKa = 11.84WTTSPGAKK282 pKa = 8.8RR283 pKa = 11.84PRR285 pKa = 11.84MSSS288 pKa = 2.83
Molecular weight: 32.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A286QV10|A0A286QV10_9GEMI Replication-associated protein OS=Maize striate mosaic virus OX=2025388 PE=3 SV=1
MM1 pKa = 7.86RR2 pKa = 11.84PMTGKK7 pKa = 10.3RR8 pKa = 11.84KK9 pKa = 8.45RR10 pKa = 11.84TAGSSSVNRR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 9.9SRR23 pKa = 11.84RR24 pKa = 11.84VLPRR28 pKa = 11.84TVAPYY33 pKa = 9.98SQLPSRR39 pKa = 11.84LPSLQVQTFASQGSAVVEE57 pKa = 4.33VKK59 pKa = 10.75KK60 pKa = 10.94GGNCLMVTSYY70 pKa = 11.4SRR72 pKa = 11.84GSDD75 pKa = 2.84EE76 pKa = 4.53SQRR79 pKa = 11.84HH80 pKa = 4.42TNEE83 pKa = 3.16TMTYY87 pKa = 10.56KK88 pKa = 10.15MSLDD92 pKa = 3.23HH93 pKa = 8.04VMVLRR98 pKa = 11.84AEE100 pKa = 4.02LCKK103 pKa = 10.82YY104 pKa = 9.98SFKK107 pKa = 10.29ATHH110 pKa = 6.59CGWVVYY116 pKa = 9.41DD117 pKa = 5.21AKK119 pKa = 10.29PTGNQVTCKK128 pKa = 10.05TIFGYY133 pKa = 9.92PDD135 pKa = 3.55GLVDD139 pKa = 6.11YY140 pKa = 7.46PTTWKK145 pKa = 10.15VARR148 pKa = 11.84DD149 pKa = 3.42VAHH152 pKa = 6.56RR153 pKa = 11.84FVVKK157 pKa = 10.51KK158 pKa = 9.24RR159 pKa = 11.84WTYY162 pKa = 10.6RR163 pKa = 11.84MEE165 pKa = 4.89SNGSNSSKK173 pKa = 10.51DD174 pKa = 3.19WSNGTGIPPCNRR186 pKa = 11.84SVYY189 pKa = 10.15VKK191 pKa = 10.57QFVSKK196 pKa = 10.29LNCRR200 pKa = 11.84TEE202 pKa = 4.0WKK204 pKa = 8.44NTTGGDD210 pKa = 3.41YY211 pKa = 11.46GDD213 pKa = 3.82IKK215 pKa = 11.12GGALYY220 pKa = 10.58VVLAAGQGMEE230 pKa = 3.86HH231 pKa = 6.38MAYY234 pKa = 8.4GTTRR238 pKa = 11.84MYY240 pKa = 10.63FKK242 pKa = 10.96SIGNQQ247 pKa = 3.0
MM1 pKa = 7.86RR2 pKa = 11.84PMTGKK7 pKa = 10.3RR8 pKa = 11.84KK9 pKa = 8.45RR10 pKa = 11.84TAGSSSVNRR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 9.9SRR23 pKa = 11.84RR24 pKa = 11.84VLPRR28 pKa = 11.84TVAPYY33 pKa = 9.98SQLPSRR39 pKa = 11.84LPSLQVQTFASQGSAVVEE57 pKa = 4.33VKK59 pKa = 10.75KK60 pKa = 10.94GGNCLMVTSYY70 pKa = 11.4SRR72 pKa = 11.84GSDD75 pKa = 2.84EE76 pKa = 4.53SQRR79 pKa = 11.84HH80 pKa = 4.42TNEE83 pKa = 3.16TMTYY87 pKa = 10.56KK88 pKa = 10.15MSLDD92 pKa = 3.23HH93 pKa = 8.04VMVLRR98 pKa = 11.84AEE100 pKa = 4.02LCKK103 pKa = 10.82YY104 pKa = 9.98SFKK107 pKa = 10.29ATHH110 pKa = 6.59CGWVVYY116 pKa = 9.41DD117 pKa = 5.21AKK119 pKa = 10.29PTGNQVTCKK128 pKa = 10.05TIFGYY133 pKa = 9.92PDD135 pKa = 3.55GLVDD139 pKa = 6.11YY140 pKa = 7.46PTTWKK145 pKa = 10.15VARR148 pKa = 11.84DD149 pKa = 3.42VAHH152 pKa = 6.56RR153 pKa = 11.84FVVKK157 pKa = 10.51KK158 pKa = 9.24RR159 pKa = 11.84WTYY162 pKa = 10.6RR163 pKa = 11.84MEE165 pKa = 4.89SNGSNSSKK173 pKa = 10.51DD174 pKa = 3.19WSNGTGIPPCNRR186 pKa = 11.84SVYY189 pKa = 10.15VKK191 pKa = 10.57QFVSKK196 pKa = 10.29LNCRR200 pKa = 11.84TEE202 pKa = 4.0WKK204 pKa = 8.44NTTGGDD210 pKa = 3.41YY211 pKa = 11.46GDD213 pKa = 3.82IKK215 pKa = 11.12GGALYY220 pKa = 10.58VVLAAGQGMEE230 pKa = 3.86HH231 pKa = 6.38MAYY234 pKa = 8.4GTTRR238 pKa = 11.84MYY240 pKa = 10.63FKK242 pKa = 10.96SIGNQQ247 pKa = 3.0
Molecular weight: 27.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
998 |
120 |
343 |
249.5 |
28.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.014 ± 0.697 | 2.605 ± 0.184 |
5.01 ± 0.598 | 4.208 ± 0.417 |
4.81 ± 0.738 | 6.613 ± 0.8 |
2.806 ± 0.605 | 4.008 ± 0.985 |
5.311 ± 0.732 | 6.112 ± 0.72 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.705 ± 0.398 | 4.008 ± 0.489 |
6.313 ± 0.742 | 3.106 ± 0.183 |
6.613 ± 0.388 | 8.116 ± 0.579 |
7.615 ± 0.56 | 6.613 ± 1.315 |
2.004 ± 0.136 | 4.409 ± 0.397 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
