
Tortoise microvirus 36
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.29
Get precalculated fractions of proteins
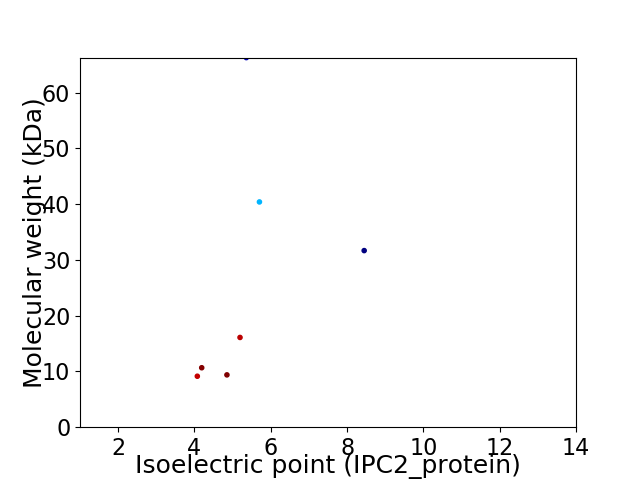
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
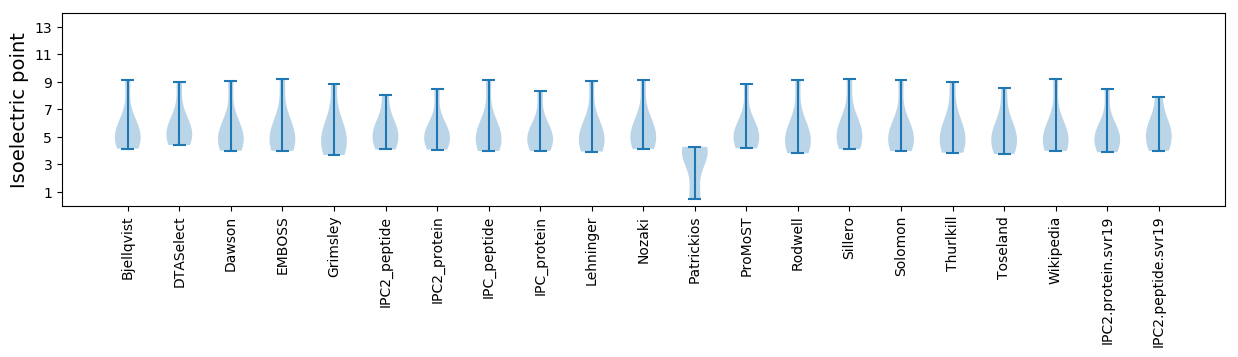
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W9M6|A0A4P8W9M6_9VIRU Uncharacterized protein OS=Tortoise microvirus 36 OX=2583139 PE=4 SV=1
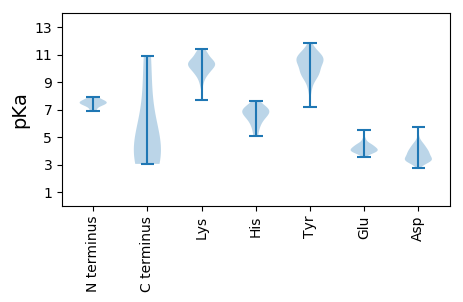
MM1 pKa = 6.89FTFHH5 pKa = 7.01SFLHH9 pKa = 6.01FLCFYY14 pKa = 10.59IGVCMFDD21 pKa = 3.41VDD23 pKa = 3.97TSDD26 pKa = 5.14GSILPSGRR34 pKa = 11.84SRR36 pKa = 11.84AVDD39 pKa = 3.35VTILFPEE46 pKa = 4.27SFIDD50 pKa = 5.75AIDD53 pKa = 3.75NLLLGDD59 pKa = 4.09TMSLPDD65 pKa = 5.63FIRR68 pKa = 11.84LACVEE73 pKa = 4.19YY74 pKa = 10.15FISTLMCC81 pKa = 4.61
MM1 pKa = 6.89FTFHH5 pKa = 7.01SFLHH9 pKa = 6.01FLCFYY14 pKa = 10.59IGVCMFDD21 pKa = 3.41VDD23 pKa = 3.97TSDD26 pKa = 5.14GSILPSGRR34 pKa = 11.84SRR36 pKa = 11.84AVDD39 pKa = 3.35VTILFPEE46 pKa = 4.27SFIDD50 pKa = 5.75AIDD53 pKa = 3.75NLLLGDD59 pKa = 4.09TMSLPDD65 pKa = 5.63FIRR68 pKa = 11.84LACVEE73 pKa = 4.19YY74 pKa = 10.15FISTLMCC81 pKa = 4.61
Molecular weight: 9.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W620|A0A4P8W620_9VIRU DNA pilot protein OS=Tortoise microvirus 36 OX=2583139 PE=4 SV=1
MM1 pKa = 7.56SCRR4 pKa = 11.84ITYY7 pKa = 9.91QSYY10 pKa = 11.24LLFCCKK16 pKa = 10.02RR17 pKa = 11.84EE18 pKa = 4.95LFDD21 pKa = 4.33CYY23 pKa = 10.61QRR25 pKa = 11.84GLGASFLTFTYY36 pKa = 11.05NDD38 pKa = 3.93DD39 pKa = 3.89CLPLYY44 pKa = 10.82GSLDD48 pKa = 3.2KK49 pKa = 11.17RR50 pKa = 11.84SFQNLLKK57 pKa = 10.46RR58 pKa = 11.84ARR60 pKa = 11.84RR61 pKa = 11.84NKK63 pKa = 9.7GLPAFKK69 pKa = 10.61YY70 pKa = 8.99LACGEE75 pKa = 4.37YY76 pKa = 10.63GDD78 pKa = 4.28KK79 pKa = 10.29FDD81 pKa = 4.09RR82 pKa = 11.84AHH84 pKa = 5.26YY85 pKa = 9.48HH86 pKa = 6.17AVFFGLTDD94 pKa = 3.3VLAKK98 pKa = 10.51QFVASHH104 pKa = 6.58WLNGYY109 pKa = 9.47VKK111 pKa = 10.6VEE113 pKa = 4.1ALRR116 pKa = 11.84SSAGLRR122 pKa = 11.84YY123 pKa = 10.1VLDD126 pKa = 3.97YY127 pKa = 10.64CCKK130 pKa = 10.57SINGDD135 pKa = 3.91LAVEE139 pKa = 4.82LYY141 pKa = 10.52DD142 pKa = 3.31NQMRR146 pKa = 11.84EE147 pKa = 4.13RR148 pKa = 11.84PFLSHH153 pKa = 5.04STRR156 pKa = 11.84MGFDD160 pKa = 2.77WLVRR164 pKa = 11.84NMDD167 pKa = 4.17EE168 pKa = 3.87IVANGFYY175 pKa = 10.43YY176 pKa = 10.51RR177 pKa = 11.84EE178 pKa = 4.22AGKK181 pKa = 8.7MLPLPKK187 pKa = 9.96YY188 pKa = 10.41FRR190 pKa = 11.84DD191 pKa = 3.33IYY193 pKa = 10.45DD194 pKa = 3.37TYY196 pKa = 10.86KK197 pKa = 10.23TFDD200 pKa = 3.15VSRR203 pKa = 11.84VLRR206 pKa = 11.84SLDD209 pKa = 3.36SHH211 pKa = 6.91AKK213 pKa = 10.22RR214 pKa = 11.84MGFPDD219 pKa = 3.33IYY221 pKa = 10.71SYY223 pKa = 11.71SDD225 pKa = 3.31YY226 pKa = 10.84ATPLKK231 pKa = 10.41EE232 pKa = 4.36KK233 pKa = 10.63EE234 pKa = 4.44FISKK238 pKa = 10.14SRR240 pKa = 11.84LAGRR244 pKa = 11.84PVDD247 pKa = 3.85DD248 pKa = 4.7SNFVSSFYY256 pKa = 10.99VPRR259 pKa = 11.84LGTNNLAKK267 pKa = 10.55SLLEE271 pKa = 3.98RR272 pKa = 4.81
MM1 pKa = 7.56SCRR4 pKa = 11.84ITYY7 pKa = 9.91QSYY10 pKa = 11.24LLFCCKK16 pKa = 10.02RR17 pKa = 11.84EE18 pKa = 4.95LFDD21 pKa = 4.33CYY23 pKa = 10.61QRR25 pKa = 11.84GLGASFLTFTYY36 pKa = 11.05NDD38 pKa = 3.93DD39 pKa = 3.89CLPLYY44 pKa = 10.82GSLDD48 pKa = 3.2KK49 pKa = 11.17RR50 pKa = 11.84SFQNLLKK57 pKa = 10.46RR58 pKa = 11.84ARR60 pKa = 11.84RR61 pKa = 11.84NKK63 pKa = 9.7GLPAFKK69 pKa = 10.61YY70 pKa = 8.99LACGEE75 pKa = 4.37YY76 pKa = 10.63GDD78 pKa = 4.28KK79 pKa = 10.29FDD81 pKa = 4.09RR82 pKa = 11.84AHH84 pKa = 5.26YY85 pKa = 9.48HH86 pKa = 6.17AVFFGLTDD94 pKa = 3.3VLAKK98 pKa = 10.51QFVASHH104 pKa = 6.58WLNGYY109 pKa = 9.47VKK111 pKa = 10.6VEE113 pKa = 4.1ALRR116 pKa = 11.84SSAGLRR122 pKa = 11.84YY123 pKa = 10.1VLDD126 pKa = 3.97YY127 pKa = 10.64CCKK130 pKa = 10.57SINGDD135 pKa = 3.91LAVEE139 pKa = 4.82LYY141 pKa = 10.52DD142 pKa = 3.31NQMRR146 pKa = 11.84EE147 pKa = 4.13RR148 pKa = 11.84PFLSHH153 pKa = 5.04STRR156 pKa = 11.84MGFDD160 pKa = 2.77WLVRR164 pKa = 11.84NMDD167 pKa = 4.17EE168 pKa = 3.87IVANGFYY175 pKa = 10.43YY176 pKa = 10.51RR177 pKa = 11.84EE178 pKa = 4.22AGKK181 pKa = 8.7MLPLPKK187 pKa = 9.96YY188 pKa = 10.41FRR190 pKa = 11.84DD191 pKa = 3.33IYY193 pKa = 10.45DD194 pKa = 3.37TYY196 pKa = 10.86KK197 pKa = 10.23TFDD200 pKa = 3.15VSRR203 pKa = 11.84VLRR206 pKa = 11.84SLDD209 pKa = 3.36SHH211 pKa = 6.91AKK213 pKa = 10.22RR214 pKa = 11.84MGFPDD219 pKa = 3.33IYY221 pKa = 10.71SYY223 pKa = 11.71SDD225 pKa = 3.31YY226 pKa = 10.84ATPLKK231 pKa = 10.41EE232 pKa = 4.36KK233 pKa = 10.63EE234 pKa = 4.44FISKK238 pKa = 10.14SRR240 pKa = 11.84LAGRR244 pKa = 11.84PVDD247 pKa = 3.85DD248 pKa = 4.7SNFVSSFYY256 pKa = 10.99VPRR259 pKa = 11.84LGTNNLAKK267 pKa = 10.55SLLEE271 pKa = 3.98RR272 pKa = 4.81
Molecular weight: 31.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1635 |
81 |
591 |
233.6 |
26.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.073 ± 1.343 | 1.774 ± 0.572 |
6.667 ± 1.091 | 4.587 ± 0.88 |
5.443 ± 0.911 | 5.26 ± 0.391 |
1.529 ± 0.237 | 5.199 ± 0.739 |
4.037 ± 1.029 | 9.235 ± 0.819 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.63 ± 0.242 | 4.771 ± 0.35 |
4.404 ± 0.766 | 3.914 ± 1.694 |
5.627 ± 0.607 | 10.398 ± 1.03 |
5.749 ± 0.697 | 4.709 ± 0.401 |
0.856 ± 0.172 | 5.138 ± 0.687 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
