
Alkalibacterium gilvum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Carnobacteriaceae; Alkalibacterium
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
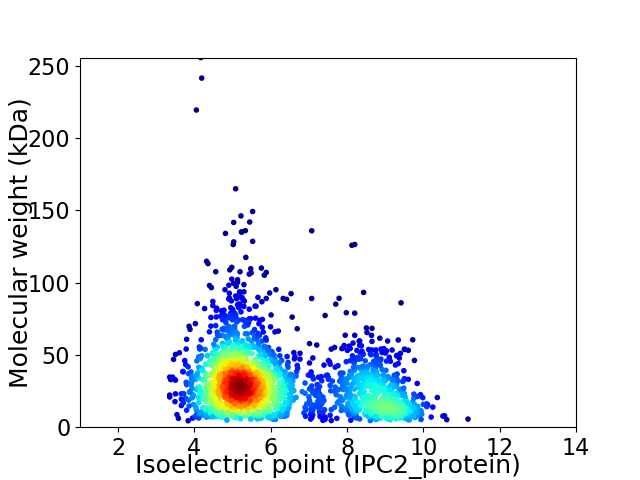
Virtual 2D-PAGE plot for 2101 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H6UL70|A0A1H6UL70_9LACT Thiol peroxidase atypical 2-Cys peroxiredoxin OS=Alkalibacterium gilvum OX=1130080 GN=SAMN04488113_13417 PE=4 SV=1
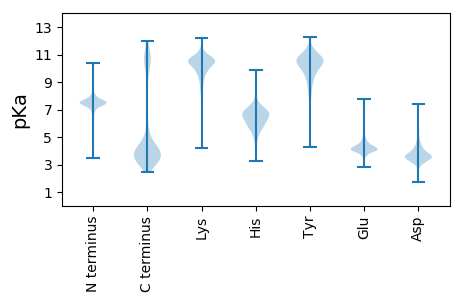
MM1 pKa = 7.54TIIIGITLFTGFKK14 pKa = 9.5TYY16 pKa = 10.63QIAQAFGSPLNRR28 pKa = 11.84NIEE31 pKa = 3.69EE32 pKa = 3.87WLYY35 pKa = 10.78IKK37 pKa = 9.85TVDD40 pKa = 3.55YY41 pKa = 11.61EE42 pKa = 4.22NFNVFDD48 pKa = 3.74QSVEE52 pKa = 4.06EE53 pKa = 4.61LFTMIDD59 pKa = 3.78EE60 pKa = 4.71EE61 pKa = 4.81VDD63 pKa = 4.03LPDD66 pKa = 4.32PLYY69 pKa = 10.4IYY71 pKa = 11.08NEE73 pKa = 3.84FDD75 pKa = 4.19LEE77 pKa = 4.12FDD79 pKa = 3.81RR80 pKa = 11.84SGQLTSFYY88 pKa = 11.07AAVTGEE94 pKa = 3.95NEE96 pKa = 3.95EE97 pKa = 5.24GVFEE101 pKa = 4.32WFLISDD107 pKa = 3.78SDD109 pKa = 4.04EE110 pKa = 4.5PNQLTINSGEE120 pKa = 4.04NPDD123 pKa = 5.24RR124 pKa = 11.84EE125 pKa = 4.31VLSNNMLLSPLFDD138 pKa = 3.85TLDD141 pKa = 3.51QLSLEE146 pKa = 4.43EE147 pKa = 5.04TIEE150 pKa = 3.85TWPAEE155 pKa = 4.28EE156 pKa = 4.43VFGIYY161 pKa = 10.03YY162 pKa = 10.41DD163 pKa = 4.97GYY165 pKa = 10.81RR166 pKa = 11.84SWGTNDD172 pKa = 2.75TGIYY176 pKa = 9.88YY177 pKa = 10.2LGNGGGPIRR186 pKa = 11.84IEE188 pKa = 5.0LYY190 pKa = 10.13DD191 pKa = 3.95QEE193 pKa = 4.29ILGYY197 pKa = 7.66TVSVYY202 pKa = 10.59VSGKK206 pKa = 8.45TEE208 pKa = 4.34TITPMRR214 pKa = 11.84YY215 pKa = 8.53IDD217 pKa = 3.51SSLNVLSEE225 pKa = 4.71PIDD228 pKa = 3.98PEE230 pKa = 4.22SEE232 pKa = 4.11KK233 pKa = 11.1PEE235 pKa = 3.5IGYY238 pKa = 9.23QVDD241 pKa = 3.67EE242 pKa = 4.66QDD244 pKa = 2.91QKK246 pKa = 11.05MYY248 pKa = 11.06YY249 pKa = 8.2LTEE252 pKa = 4.52EE253 pKa = 3.27IGYY256 pKa = 9.91RR257 pKa = 11.84LAVLDD262 pKa = 4.28AATGSRR268 pKa = 11.84WYY270 pKa = 10.62GLEE273 pKa = 3.75KK274 pKa = 10.55TEE276 pKa = 5.09DD277 pKa = 3.61AGEE280 pKa = 4.2SWVSVNPDD288 pKa = 3.39PFDD291 pKa = 3.73GRR293 pKa = 11.84SGSSTGLKK301 pKa = 10.08FFNEE305 pKa = 4.65DD306 pKa = 3.18FGFMAIARR314 pKa = 11.84RR315 pKa = 11.84DD316 pKa = 3.4QATLFRR322 pKa = 11.84TEE324 pKa = 3.95NGGEE328 pKa = 4.05IISPVTFPDD337 pKa = 3.42VQVPLIDD344 pKa = 4.93DD345 pKa = 3.42EE346 pKa = 4.95TYY348 pKa = 11.09NPFRR352 pKa = 11.84FPDD355 pKa = 3.46MPYY358 pKa = 10.65EE359 pKa = 4.36EE360 pKa = 5.11NDD362 pKa = 3.28EE363 pKa = 4.87LYY365 pKa = 9.61VTVGQDD371 pKa = 3.18PNGDD375 pKa = 3.81YY376 pKa = 11.38NSGAKK381 pKa = 9.89ALYY384 pKa = 10.25KK385 pKa = 10.69SVDD388 pKa = 3.82DD389 pKa = 4.54GKK391 pKa = 8.28TWTYY395 pKa = 10.23VDD397 pKa = 3.56EE398 pKa = 4.55VGIGFGNDD406 pKa = 2.65
MM1 pKa = 7.54TIIIGITLFTGFKK14 pKa = 9.5TYY16 pKa = 10.63QIAQAFGSPLNRR28 pKa = 11.84NIEE31 pKa = 3.69EE32 pKa = 3.87WLYY35 pKa = 10.78IKK37 pKa = 9.85TVDD40 pKa = 3.55YY41 pKa = 11.61EE42 pKa = 4.22NFNVFDD48 pKa = 3.74QSVEE52 pKa = 4.06EE53 pKa = 4.61LFTMIDD59 pKa = 3.78EE60 pKa = 4.71EE61 pKa = 4.81VDD63 pKa = 4.03LPDD66 pKa = 4.32PLYY69 pKa = 10.4IYY71 pKa = 11.08NEE73 pKa = 3.84FDD75 pKa = 4.19LEE77 pKa = 4.12FDD79 pKa = 3.81RR80 pKa = 11.84SGQLTSFYY88 pKa = 11.07AAVTGEE94 pKa = 3.95NEE96 pKa = 3.95EE97 pKa = 5.24GVFEE101 pKa = 4.32WFLISDD107 pKa = 3.78SDD109 pKa = 4.04EE110 pKa = 4.5PNQLTINSGEE120 pKa = 4.04NPDD123 pKa = 5.24RR124 pKa = 11.84EE125 pKa = 4.31VLSNNMLLSPLFDD138 pKa = 3.85TLDD141 pKa = 3.51QLSLEE146 pKa = 4.43EE147 pKa = 5.04TIEE150 pKa = 3.85TWPAEE155 pKa = 4.28EE156 pKa = 4.43VFGIYY161 pKa = 10.03YY162 pKa = 10.41DD163 pKa = 4.97GYY165 pKa = 10.81RR166 pKa = 11.84SWGTNDD172 pKa = 2.75TGIYY176 pKa = 9.88YY177 pKa = 10.2LGNGGGPIRR186 pKa = 11.84IEE188 pKa = 5.0LYY190 pKa = 10.13DD191 pKa = 3.95QEE193 pKa = 4.29ILGYY197 pKa = 7.66TVSVYY202 pKa = 10.59VSGKK206 pKa = 8.45TEE208 pKa = 4.34TITPMRR214 pKa = 11.84YY215 pKa = 8.53IDD217 pKa = 3.51SSLNVLSEE225 pKa = 4.71PIDD228 pKa = 3.98PEE230 pKa = 4.22SEE232 pKa = 4.11KK233 pKa = 11.1PEE235 pKa = 3.5IGYY238 pKa = 9.23QVDD241 pKa = 3.67EE242 pKa = 4.66QDD244 pKa = 2.91QKK246 pKa = 11.05MYY248 pKa = 11.06YY249 pKa = 8.2LTEE252 pKa = 4.52EE253 pKa = 3.27IGYY256 pKa = 9.91RR257 pKa = 11.84LAVLDD262 pKa = 4.28AATGSRR268 pKa = 11.84WYY270 pKa = 10.62GLEE273 pKa = 3.75KK274 pKa = 10.55TEE276 pKa = 5.09DD277 pKa = 3.61AGEE280 pKa = 4.2SWVSVNPDD288 pKa = 3.39PFDD291 pKa = 3.73GRR293 pKa = 11.84SGSSTGLKK301 pKa = 10.08FFNEE305 pKa = 4.65DD306 pKa = 3.18FGFMAIARR314 pKa = 11.84RR315 pKa = 11.84DD316 pKa = 3.4QATLFRR322 pKa = 11.84TEE324 pKa = 3.95NGGEE328 pKa = 4.05IISPVTFPDD337 pKa = 3.42VQVPLIDD344 pKa = 4.93DD345 pKa = 3.42EE346 pKa = 4.95TYY348 pKa = 11.09NPFRR352 pKa = 11.84FPDD355 pKa = 3.46MPYY358 pKa = 10.65EE359 pKa = 4.36EE360 pKa = 5.11NDD362 pKa = 3.28EE363 pKa = 4.87LYY365 pKa = 9.61VTVGQDD371 pKa = 3.18PNGDD375 pKa = 3.81YY376 pKa = 11.38NSGAKK381 pKa = 9.89ALYY384 pKa = 10.25KK385 pKa = 10.69SVDD388 pKa = 3.82DD389 pKa = 4.54GKK391 pKa = 8.28TWTYY395 pKa = 10.23VDD397 pKa = 3.56EE398 pKa = 4.55VGIGFGNDD406 pKa = 2.65
Molecular weight: 46.09 kDa
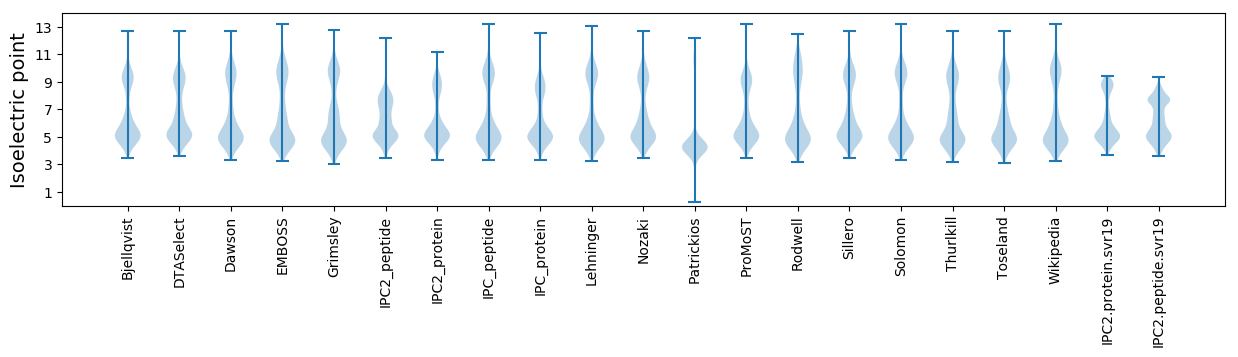
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H6SKJ8|A0A1H6SKJ8_9LACT Transcriptional regulator OS=Alkalibacterium gilvum OX=1130080 GN=SAMN04488113_10714 PE=4 SV=1
MM1 pKa = 7.35AQGLTYY7 pKa = 10.18QPKK10 pKa = 8.23KK11 pKa = 10.01RR12 pKa = 11.84KK13 pKa = 7.66RR14 pKa = 11.84QRR16 pKa = 11.84VHH18 pKa = 6.34GFRR21 pKa = 11.84KK22 pKa = 10.04RR23 pKa = 11.84MSTKK27 pKa = 9.72NGRR30 pKa = 11.84RR31 pKa = 11.84VLKK34 pKa = 9.89NRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 8.47GRR41 pKa = 11.84KK42 pKa = 9.15RR43 pKa = 11.84ISAA46 pKa = 3.72
MM1 pKa = 7.35AQGLTYY7 pKa = 10.18QPKK10 pKa = 8.23KK11 pKa = 10.01RR12 pKa = 11.84KK13 pKa = 7.66RR14 pKa = 11.84QRR16 pKa = 11.84VHH18 pKa = 6.34GFRR21 pKa = 11.84KK22 pKa = 10.04RR23 pKa = 11.84MSTKK27 pKa = 9.72NGRR30 pKa = 11.84RR31 pKa = 11.84VLKK34 pKa = 9.89NRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 8.47GRR41 pKa = 11.84KK42 pKa = 9.15RR43 pKa = 11.84ISAA46 pKa = 3.72
Molecular weight: 5.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
615918 |
39 |
2328 |
293.2 |
33.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.373 ± 0.051 | 0.441 ± 0.012 |
5.93 ± 0.053 | 7.975 ± 0.071 |
4.419 ± 0.05 | 6.316 ± 0.045 |
1.895 ± 0.023 | 7.849 ± 0.059 |
7.496 ± 0.053 | 9.607 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.71 ± 0.024 | 5.049 ± 0.046 |
3.202 ± 0.025 | 3.428 ± 0.03 |
3.918 ± 0.04 | 6.332 ± 0.036 |
5.744 ± 0.035 | 6.689 ± 0.04 |
0.859 ± 0.019 | 3.768 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
