
Cucumber necrosis virus (CNV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Tombusvirus
Average proteome isoelectric point is 8.43
Get precalculated fractions of proteins
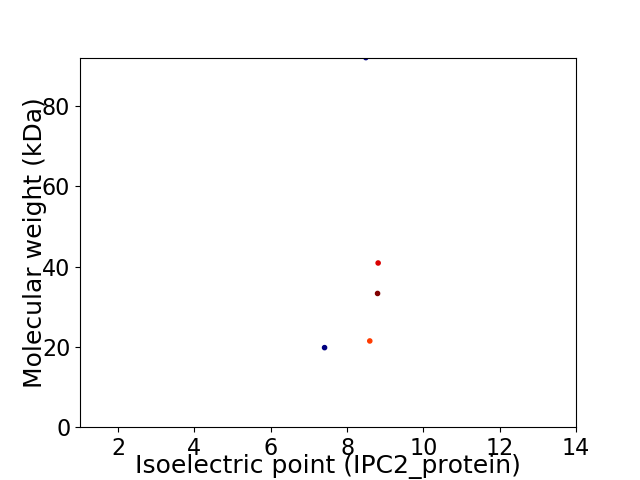
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P15185|MVP_CNV Movement protein OS=Cucumber necrosis virus OX=12143 GN=ORF3 PE=3 SV=1
MM1 pKa = 7.3EE2 pKa = 5.06RR3 pKa = 11.84AIQRR7 pKa = 11.84SDD9 pKa = 3.0ARR11 pKa = 11.84EE12 pKa = 3.73QANSEE17 pKa = 4.07RR18 pKa = 11.84WDD20 pKa = 3.7GRR22 pKa = 11.84CGGTITPFKK31 pKa = 10.79LPDD34 pKa = 3.67EE35 pKa = 4.76SPSLLEE41 pKa = 3.9WRR43 pKa = 11.84LHH45 pKa = 5.33NSEE48 pKa = 4.05EE49 pKa = 5.04SEE51 pKa = 5.1DD52 pKa = 3.73KK53 pKa = 11.04DD54 pKa = 3.5HH55 pKa = 7.42PLGFKK60 pKa = 10.23EE61 pKa = 4.04SWSFGKK67 pKa = 10.55VVFKK71 pKa = 10.76RR72 pKa = 11.84YY73 pKa = 9.63LRR75 pKa = 11.84YY76 pKa = 10.52DD77 pKa = 3.29GTEE80 pKa = 3.89TSLHH84 pKa = 5.47RR85 pKa = 11.84TLGSWEE91 pKa = 4.02RR92 pKa = 11.84NSVNDD97 pKa = 3.41AASRR101 pKa = 11.84FLGVSQIGCTYY112 pKa = 10.29SIRR115 pKa = 11.84FRR117 pKa = 11.84GSCLTLSGGSRR128 pKa = 11.84TLQRR132 pKa = 11.84LIEE135 pKa = 3.89MAIRR139 pKa = 11.84TKK141 pKa = 10.3RR142 pKa = 11.84TMLQLTPCEE151 pKa = 4.17VEE153 pKa = 4.31GNVSRR158 pKa = 11.84RR159 pKa = 11.84RR160 pKa = 11.84PQGSEE165 pKa = 3.35AFEE168 pKa = 4.32NKK170 pKa = 9.68EE171 pKa = 4.03SEE173 pKa = 4.2
MM1 pKa = 7.3EE2 pKa = 5.06RR3 pKa = 11.84AIQRR7 pKa = 11.84SDD9 pKa = 3.0ARR11 pKa = 11.84EE12 pKa = 3.73QANSEE17 pKa = 4.07RR18 pKa = 11.84WDD20 pKa = 3.7GRR22 pKa = 11.84CGGTITPFKK31 pKa = 10.79LPDD34 pKa = 3.67EE35 pKa = 4.76SPSLLEE41 pKa = 3.9WRR43 pKa = 11.84LHH45 pKa = 5.33NSEE48 pKa = 4.05EE49 pKa = 5.04SEE51 pKa = 5.1DD52 pKa = 3.73KK53 pKa = 11.04DD54 pKa = 3.5HH55 pKa = 7.42PLGFKK60 pKa = 10.23EE61 pKa = 4.04SWSFGKK67 pKa = 10.55VVFKK71 pKa = 10.76RR72 pKa = 11.84YY73 pKa = 9.63LRR75 pKa = 11.84YY76 pKa = 10.52DD77 pKa = 3.29GTEE80 pKa = 3.89TSLHH84 pKa = 5.47RR85 pKa = 11.84TLGSWEE91 pKa = 4.02RR92 pKa = 11.84NSVNDD97 pKa = 3.41AASRR101 pKa = 11.84FLGVSQIGCTYY112 pKa = 10.29SIRR115 pKa = 11.84FRR117 pKa = 11.84GSCLTLSGGSRR128 pKa = 11.84TLQRR132 pKa = 11.84LIEE135 pKa = 3.89MAIRR139 pKa = 11.84TKK141 pKa = 10.3RR142 pKa = 11.84TMLQLTPCEE151 pKa = 4.17VEE153 pKa = 4.31GNVSRR158 pKa = 11.84RR159 pKa = 11.84RR160 pKa = 11.84PQGSEE165 pKa = 3.35AFEE168 pKa = 4.32NKK170 pKa = 9.68EE171 pKa = 4.03SEE173 pKa = 4.2
Molecular weight: 19.81 kDa
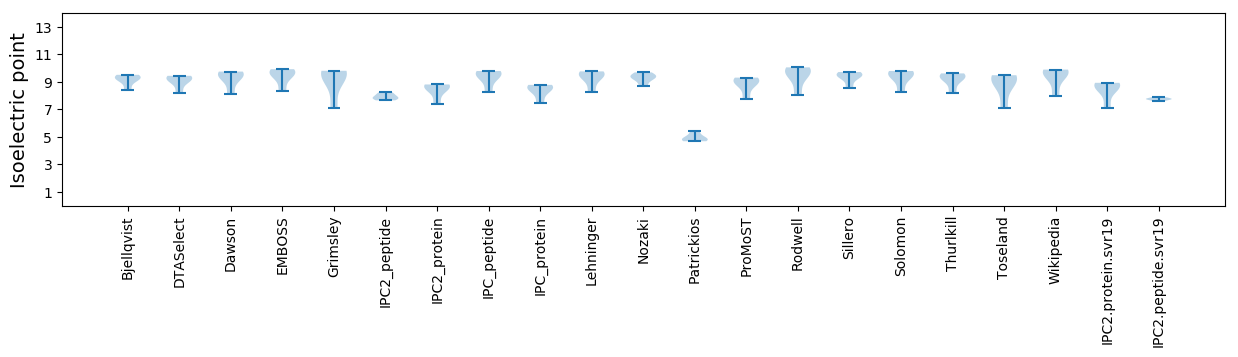
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q66164|Q66164_CNV RNA-directed RNA polymerase OS=Cucumber necrosis virus OX=12143 PE=4 SV=1
MM1 pKa = 7.5DD2 pKa = 5.19TIKK5 pKa = 10.95RR6 pKa = 11.84MLWPKK11 pKa = 10.64KK12 pKa = 9.66EE13 pKa = 3.93IFVGTFATGVEE24 pKa = 4.43RR25 pKa = 11.84DD26 pKa = 3.42TSVDD30 pKa = 2.92IFQLVCRR37 pKa = 11.84VVLRR41 pKa = 11.84YY42 pKa = 8.72MRR44 pKa = 11.84TGKK47 pKa = 9.8IEE49 pKa = 4.19NNTDD53 pKa = 3.04SLGNFIVEE61 pKa = 4.63LLKK64 pKa = 10.18TDD66 pKa = 5.21CAAKK70 pKa = 10.03WEE72 pKa = 4.24WFMKK76 pKa = 9.58RR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84VGDD82 pKa = 3.54YY83 pKa = 10.84AKK85 pKa = 10.53SLAIASIPVIPLLSYY100 pKa = 10.2ATMKK104 pKa = 9.63KK105 pKa = 8.2TVALRR110 pKa = 11.84AFGNEE115 pKa = 3.24LSFNIRR121 pKa = 11.84VPRR124 pKa = 11.84PSVPKK129 pKa = 10.39KK130 pKa = 10.83GLLLRR135 pKa = 11.84LAAGLALAPICALAMYY151 pKa = 7.64ATLPRR156 pKa = 11.84EE157 pKa = 3.87KK158 pKa = 10.77LSVFKK163 pKa = 11.04LRR165 pKa = 11.84TEE167 pKa = 4.26ARR169 pKa = 11.84AHH171 pKa = 5.9MEE173 pKa = 4.18DD174 pKa = 3.5EE175 pKa = 5.34RR176 pKa = 11.84EE177 pKa = 4.2ATDD180 pKa = 3.71CLVVEE185 pKa = 5.21PARR188 pKa = 11.84EE189 pKa = 3.99LKK191 pKa = 10.99GKK193 pKa = 10.26DD194 pKa = 3.6GEE196 pKa = 4.5DD197 pKa = 3.55LLTGSRR203 pKa = 11.84MTKK206 pKa = 10.3VIASTGRR213 pKa = 11.84PRR215 pKa = 11.84RR216 pKa = 11.84RR217 pKa = 11.84PYY219 pKa = 9.67AAKK222 pKa = 9.43IAQVARR228 pKa = 11.84AKK230 pKa = 10.52VGYY233 pKa = 10.07LKK235 pKa = 9.25NTPEE239 pKa = 3.78NRR241 pKa = 11.84LIYY244 pKa = 9.39QRR246 pKa = 11.84VMIEE250 pKa = 5.34IMDD253 pKa = 4.32KK254 pKa = 11.03DD255 pKa = 3.68CVRR258 pKa = 11.84YY259 pKa = 9.97VDD261 pKa = 4.33RR262 pKa = 11.84DD263 pKa = 3.8VILPLAIGCCFVYY276 pKa = 10.38PDD278 pKa = 4.17GVEE281 pKa = 3.91EE282 pKa = 5.03SAALWGSDD290 pKa = 2.76EE291 pKa = 4.24SLGVKK296 pKa = 9.67
MM1 pKa = 7.5DD2 pKa = 5.19TIKK5 pKa = 10.95RR6 pKa = 11.84MLWPKK11 pKa = 10.64KK12 pKa = 9.66EE13 pKa = 3.93IFVGTFATGVEE24 pKa = 4.43RR25 pKa = 11.84DD26 pKa = 3.42TSVDD30 pKa = 2.92IFQLVCRR37 pKa = 11.84VVLRR41 pKa = 11.84YY42 pKa = 8.72MRR44 pKa = 11.84TGKK47 pKa = 9.8IEE49 pKa = 4.19NNTDD53 pKa = 3.04SLGNFIVEE61 pKa = 4.63LLKK64 pKa = 10.18TDD66 pKa = 5.21CAAKK70 pKa = 10.03WEE72 pKa = 4.24WFMKK76 pKa = 9.58RR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84VGDD82 pKa = 3.54YY83 pKa = 10.84AKK85 pKa = 10.53SLAIASIPVIPLLSYY100 pKa = 10.2ATMKK104 pKa = 9.63KK105 pKa = 8.2TVALRR110 pKa = 11.84AFGNEE115 pKa = 3.24LSFNIRR121 pKa = 11.84VPRR124 pKa = 11.84PSVPKK129 pKa = 10.39KK130 pKa = 10.83GLLLRR135 pKa = 11.84LAAGLALAPICALAMYY151 pKa = 7.64ATLPRR156 pKa = 11.84EE157 pKa = 3.87KK158 pKa = 10.77LSVFKK163 pKa = 11.04LRR165 pKa = 11.84TEE167 pKa = 4.26ARR169 pKa = 11.84AHH171 pKa = 5.9MEE173 pKa = 4.18DD174 pKa = 3.5EE175 pKa = 5.34RR176 pKa = 11.84EE177 pKa = 4.2ATDD180 pKa = 3.71CLVVEE185 pKa = 5.21PARR188 pKa = 11.84EE189 pKa = 3.99LKK191 pKa = 10.99GKK193 pKa = 10.26DD194 pKa = 3.6GEE196 pKa = 4.5DD197 pKa = 3.55LLTGSRR203 pKa = 11.84MTKK206 pKa = 10.3VIASTGRR213 pKa = 11.84PRR215 pKa = 11.84RR216 pKa = 11.84RR217 pKa = 11.84PYY219 pKa = 9.67AAKK222 pKa = 9.43IAQVARR228 pKa = 11.84AKK230 pKa = 10.52VGYY233 pKa = 10.07LKK235 pKa = 9.25NTPEE239 pKa = 3.78NRR241 pKa = 11.84LIYY244 pKa = 9.39QRR246 pKa = 11.84VMIEE250 pKa = 5.34IMDD253 pKa = 4.32KK254 pKa = 11.03DD255 pKa = 3.68CVRR258 pKa = 11.84YY259 pKa = 9.97VDD261 pKa = 4.33RR262 pKa = 11.84DD263 pKa = 3.8VILPLAIGCCFVYY276 pKa = 10.38PDD278 pKa = 4.17GVEE281 pKa = 3.91EE282 pKa = 5.03SAALWGSDD290 pKa = 2.76EE291 pKa = 4.24SLGVKK296 pKa = 9.67
Molecular weight: 33.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1857 |
173 |
818 |
371.4 |
41.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.916 ± 0.836 | 1.723 ± 0.453 |
4.793 ± 0.302 | 6.031 ± 0.744 |
3.877 ± 0.315 | 7.324 ± 0.481 |
1.454 ± 0.409 | 4.685 ± 0.331 |
6.031 ± 0.637 | 9.639 ± 0.509 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.262 ± 0.418 | 4.093 ± 0.873 |
4.847 ± 0.253 | 2.423 ± 0.33 |
7.647 ± 0.933 | 6.462 ± 1.121 |
5.924 ± 0.485 | 8.024 ± 0.575 |
1.562 ± 0.133 | 3.231 ± 0.228 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
