
Halococcus hamelinensis 100A6
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Halococcaceae; Halococcus; Halococcus hamelinensis
Average proteome isoelectric point is 5.0
Get precalculated fractions of proteins
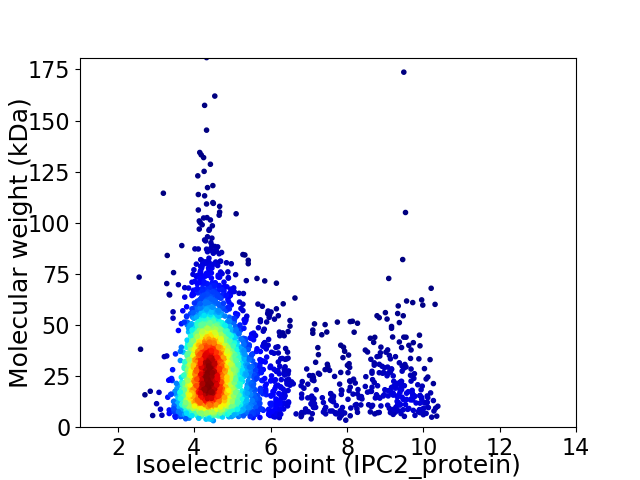
Virtual 2D-PAGE plot for 3400 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
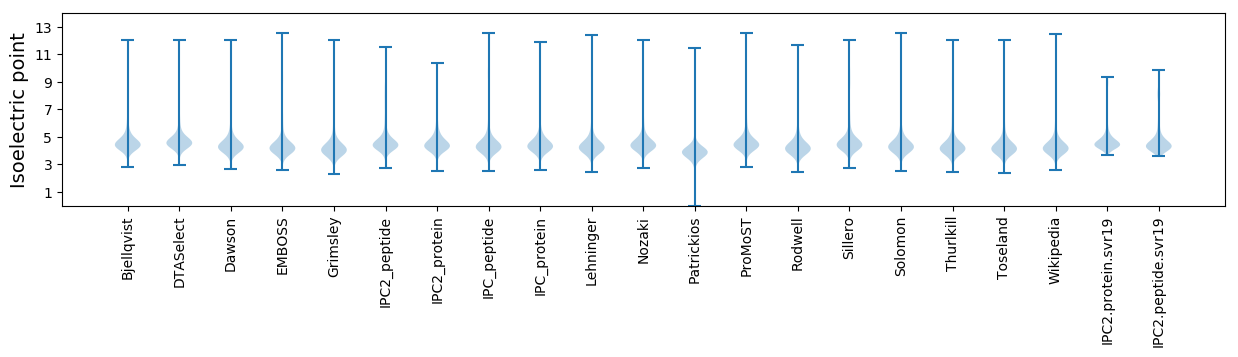
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M0M748|M0M748_9EURY Na+/H+ antiporter OS=Halococcus hamelinensis 100A6 OX=1132509 GN=C447_04717 PE=4 SV=1
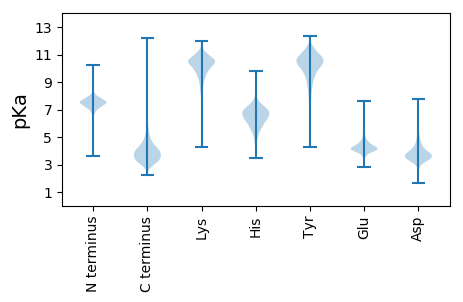
MM1 pKa = 6.93TTTAPDD7 pKa = 3.33RR8 pKa = 11.84FGGALCPTITPFADD22 pKa = 4.0DD23 pKa = 3.87EE24 pKa = 4.82VSTAGLEE31 pKa = 4.07TLIEE35 pKa = 4.12HH36 pKa = 6.43VIEE39 pKa = 4.69GGIEE43 pKa = 4.01GLVPCGTTGEE53 pKa = 4.24FASLSTTEE61 pKa = 3.83YY62 pKa = 8.81RR63 pKa = 11.84TVIEE67 pKa = 4.37TTVDD71 pKa = 3.35VADD74 pKa = 3.81GRR76 pKa = 11.84VPVMAGTAGTSVAGTVEE93 pKa = 4.38NIAFAAEE100 pKa = 3.96AGADD104 pKa = 3.41AALITLPYY112 pKa = 9.95FHH114 pKa = 7.05TANDD118 pKa = 3.6EE119 pKa = 4.35AGNEE123 pKa = 3.96AFITAVADD131 pKa = 4.0EE132 pKa = 4.76AALPCYY138 pKa = 10.06LYY140 pKa = 10.52NIPSCTGAPIAPEE153 pKa = 4.17TVSTVAGHH161 pKa = 6.94DD162 pKa = 3.72SVVGLKK168 pKa = 10.75DD169 pKa = 3.17SGGDD173 pKa = 3.21FTYY176 pKa = 10.22FQDD179 pKa = 3.54IVEE182 pKa = 4.28RR183 pKa = 11.84TPADD187 pKa = 3.52FQVYY191 pKa = 9.41PGSDD195 pKa = 3.2SLLAWGLLGGAAGGVCALSNVVPEE219 pKa = 4.59AFAEE223 pKa = 4.13LVAAVEE229 pKa = 4.27RR230 pKa = 11.84GDD232 pKa = 4.26IEE234 pKa = 4.46TATDD238 pKa = 3.87LQAGITALFQACVAHH253 pKa = 6.73GFAPTAKK260 pKa = 10.17AGLAVRR266 pKa = 11.84GVLPEE271 pKa = 3.87ATVRR275 pKa = 11.84PPLVEE280 pKa = 4.13LDD282 pKa = 3.09GDD284 pKa = 3.59ARR286 pKa = 11.84DD287 pKa = 3.95AVEE290 pKa = 3.9TAIDD294 pKa = 3.79DD295 pKa = 3.73VLTAVEE301 pKa = 4.61ASAA304 pKa = 4.93
MM1 pKa = 6.93TTTAPDD7 pKa = 3.33RR8 pKa = 11.84FGGALCPTITPFADD22 pKa = 4.0DD23 pKa = 3.87EE24 pKa = 4.82VSTAGLEE31 pKa = 4.07TLIEE35 pKa = 4.12HH36 pKa = 6.43VIEE39 pKa = 4.69GGIEE43 pKa = 4.01GLVPCGTTGEE53 pKa = 4.24FASLSTTEE61 pKa = 3.83YY62 pKa = 8.81RR63 pKa = 11.84TVIEE67 pKa = 4.37TTVDD71 pKa = 3.35VADD74 pKa = 3.81GRR76 pKa = 11.84VPVMAGTAGTSVAGTVEE93 pKa = 4.38NIAFAAEE100 pKa = 3.96AGADD104 pKa = 3.41AALITLPYY112 pKa = 9.95FHH114 pKa = 7.05TANDD118 pKa = 3.6EE119 pKa = 4.35AGNEE123 pKa = 3.96AFITAVADD131 pKa = 4.0EE132 pKa = 4.76AALPCYY138 pKa = 10.06LYY140 pKa = 10.52NIPSCTGAPIAPEE153 pKa = 4.17TVSTVAGHH161 pKa = 6.94DD162 pKa = 3.72SVVGLKK168 pKa = 10.75DD169 pKa = 3.17SGGDD173 pKa = 3.21FTYY176 pKa = 10.22FQDD179 pKa = 3.54IVEE182 pKa = 4.28RR183 pKa = 11.84TPADD187 pKa = 3.52FQVYY191 pKa = 9.41PGSDD195 pKa = 3.2SLLAWGLLGGAAGGVCALSNVVPEE219 pKa = 4.59AFAEE223 pKa = 4.13LVAAVEE229 pKa = 4.27RR230 pKa = 11.84GDD232 pKa = 4.26IEE234 pKa = 4.46TATDD238 pKa = 3.87LQAGITALFQACVAHH253 pKa = 6.73GFAPTAKK260 pKa = 10.17AGLAVRR266 pKa = 11.84GVLPEE271 pKa = 3.87ATVRR275 pKa = 11.84PPLVEE280 pKa = 4.13LDD282 pKa = 3.09GDD284 pKa = 3.59ARR286 pKa = 11.84DD287 pKa = 3.95AVEE290 pKa = 3.9TAIDD294 pKa = 3.79DD295 pKa = 3.73VLTAVEE301 pKa = 4.61ASAA304 pKa = 4.93
Molecular weight: 30.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M0LWE5|M0LWE5_9EURY Uncharacterized protein OS=Halococcus hamelinensis 100A6 OX=1132509 GN=C447_12485 PE=4 SV=1
MM1 pKa = 7.56GARR4 pKa = 11.84SRR6 pKa = 11.84WPCVIHH12 pKa = 7.12RR13 pKa = 11.84FHH15 pKa = 7.1KK16 pKa = 10.25NARR19 pKa = 11.84KK20 pKa = 9.53RR21 pKa = 11.84LRR23 pKa = 11.84EE24 pKa = 4.09LEE26 pKa = 3.98TRR28 pKa = 11.84PLIKK32 pKa = 10.4HH33 pKa = 6.08CVCAPYY39 pKa = 10.88DD40 pKa = 3.53HH41 pKa = 6.99AHH43 pKa = 6.55NARR46 pKa = 11.84IDD48 pKa = 3.65EE49 pKa = 4.97DD50 pKa = 4.03IYY52 pKa = 11.16AQRR55 pKa = 11.84SMTEE59 pKa = 4.2TVNSAMKK66 pKa = 10.28RR67 pKa = 11.84SLGYY71 pKa = 9.22VVRR74 pKa = 11.84ACDD77 pKa = 2.9WWRR80 pKa = 11.84VPRR83 pKa = 11.84DD84 pKa = 3.36RR85 pKa = 11.84SDD87 pKa = 3.64VYY89 pKa = 11.02CLQHH93 pKa = 4.48QTLRR97 pKa = 11.84HH98 pKa = 4.63TVKK101 pKa = 9.71STALQRR107 pKa = 11.84FNRR110 pKa = 11.84AKK112 pKa = 9.23QTT114 pKa = 3.42
MM1 pKa = 7.56GARR4 pKa = 11.84SRR6 pKa = 11.84WPCVIHH12 pKa = 7.12RR13 pKa = 11.84FHH15 pKa = 7.1KK16 pKa = 10.25NARR19 pKa = 11.84KK20 pKa = 9.53RR21 pKa = 11.84LRR23 pKa = 11.84EE24 pKa = 4.09LEE26 pKa = 3.98TRR28 pKa = 11.84PLIKK32 pKa = 10.4HH33 pKa = 6.08CVCAPYY39 pKa = 10.88DD40 pKa = 3.53HH41 pKa = 6.99AHH43 pKa = 6.55NARR46 pKa = 11.84IDD48 pKa = 3.65EE49 pKa = 4.97DD50 pKa = 4.03IYY52 pKa = 11.16AQRR55 pKa = 11.84SMTEE59 pKa = 4.2TVNSAMKK66 pKa = 10.28RR67 pKa = 11.84SLGYY71 pKa = 9.22VVRR74 pKa = 11.84ACDD77 pKa = 2.9WWRR80 pKa = 11.84VPRR83 pKa = 11.84DD84 pKa = 3.36RR85 pKa = 11.84SDD87 pKa = 3.64VYY89 pKa = 11.02CLQHH93 pKa = 4.48QTLRR97 pKa = 11.84HH98 pKa = 4.63TVKK101 pKa = 9.71STALQRR107 pKa = 11.84FNRR110 pKa = 11.84AKK112 pKa = 9.23QTT114 pKa = 3.42
Molecular weight: 13.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
957097 |
29 |
1650 |
281.5 |
30.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.905 ± 0.059 | 0.731 ± 0.013 |
7.95 ± 0.056 | 8.171 ± 0.061 |
3.503 ± 0.031 | 8.906 ± 0.045 |
2.025 ± 0.021 | 3.925 ± 0.034 |
1.668 ± 0.027 | 9.084 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.802 ± 0.018 | 2.436 ± 0.026 |
4.698 ± 0.032 | 2.227 ± 0.028 |
6.658 ± 0.047 | 5.577 ± 0.038 |
6.574 ± 0.036 | 9.299 ± 0.051 |
1.165 ± 0.018 | 2.691 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
